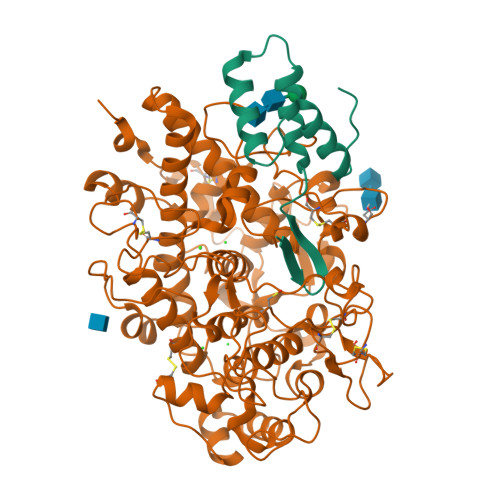
Immune evasion by a staphylococcal inhibitor of myeloperoxidase.
de Jong, N.W.M., Ramyar, K.X., Guerra, F.E., Nijland, R., Fevre, C., Voyich, J.M., McCarthy, A.J., Garcia, B.L., van Kessel, K.P.M., van Strijp, J.A.G., Geisbrecht, B.V., Haas, P.A.(2017) Proc Natl Acad Sci U S A 114: 9439-9444
- PubMed: 28808028
- DOI: https://doi.org/10.1073/pnas.1707032114
- Primary Citation of Related Structures:
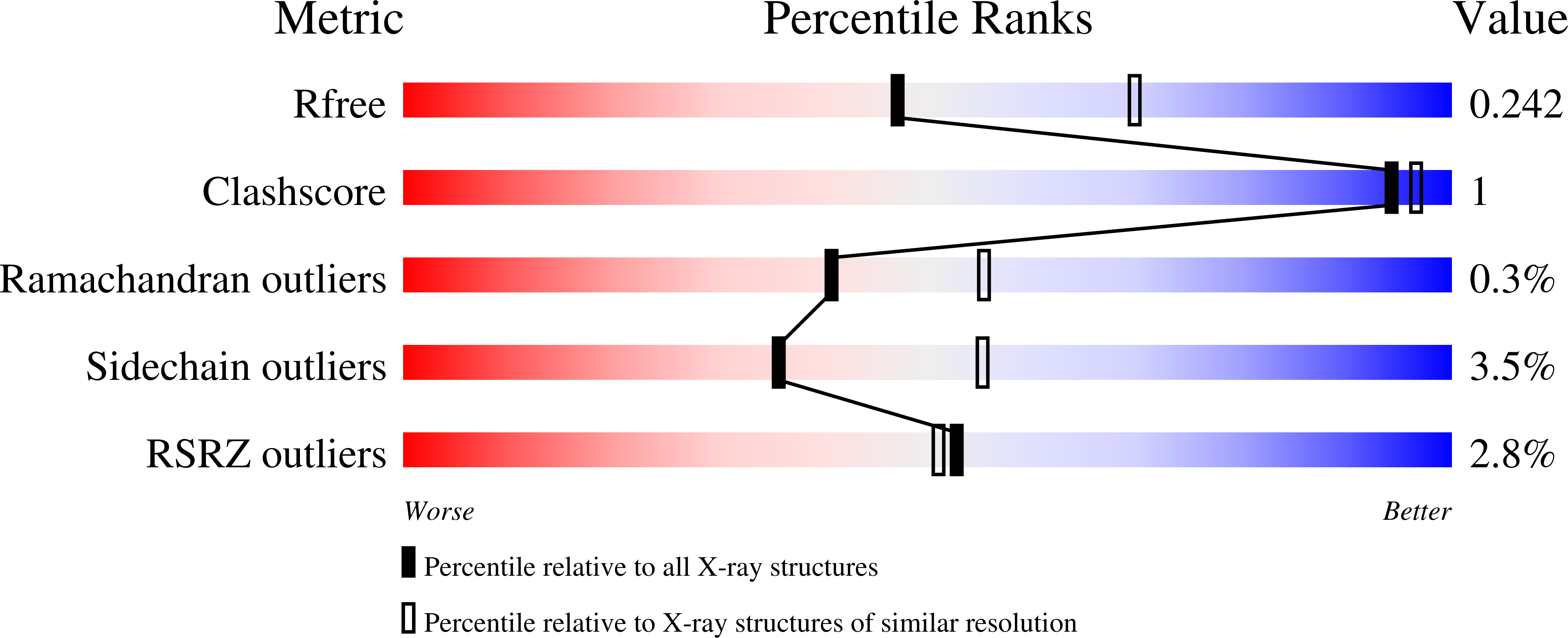
5UZU - PubMed Abstract:
Staphylococcus aureus is highly adapted to its host and has evolved many strategies to resist opsonization and phagocytosis. Even after uptake by neutrophils, S. aureus shows resistance to killing, which suggests the presence of phagosomal immune evasion molecules. With the aid of secretome phage display, we identified a highly conserved protein that specifically binds and inhibits human myeloperoxidase (MPO), a major player in the oxidative defense of neutrophils. We have named this protein "staphylococcal peroxidase inhibitor" (SPIN). To gain insight into inhibition of MPO by SPIN, we solved the cocrystal structure of SPIN bound to a recombinant form of human MPO at 2.4-Å resolution. This structure reveals that SPIN acts as a molecular plug that prevents H 2 O 2 substrate access to the MPO active site. In subsequent experiments, we observed that SPIN expression increases inside the neutrophil phagosome, where MPO is located, compared with outside the neutrophil. Moreover, bacteria with a deleted gene encoding SPIN showed decreased survival compared with WT bacteria after phagocytosis by neutrophils. Taken together, our results demonstrate that S. aureus secretes a unique proteinaceous MPO inhibitor to enhance survival by interfering with MPO-mediated killing.
Organizational Affiliation:
Medical Microbiology, University Medical Center Utrecht, 3584 CX Utrecht, The Netherlands.