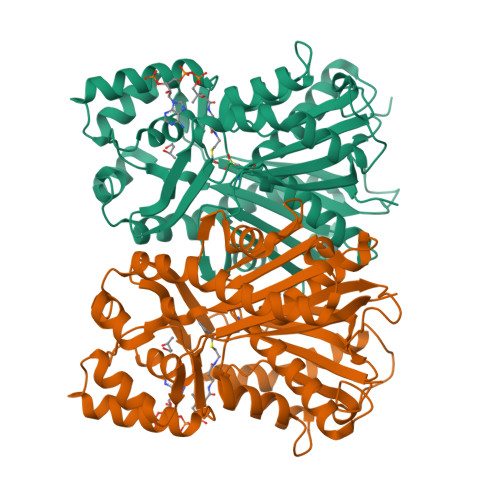
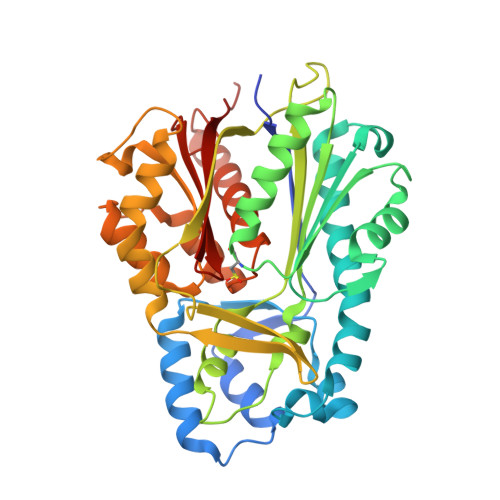
Molecular architectures of benzoic acid-specific type III polyketide synthases.
Stewart, C., Woods, K., Macias, G., Allan, A.C., Hellens, R.P., Noel, J.P.(2017) Acta Crystallogr D Struct Biol 73: 1007-1019
- PubMed: 29199980
- DOI: https://doi.org/10.1107/S2059798317016618
- Primary Citation of Related Structures:
5UC5, 5UCO, 5W8Q, 5WC4 - PubMed Abstract:
Biphenyl synthase and benzophenone synthase constitute an evolutionarily distinct clade of type III polyketide synthases (PKSs) that use benzoic acid-derived substrates to produce defense metabolites in plants. The use of benzoyl-CoA as an endogenous substrate is unusual for type III PKSs. Moreover, sequence analyses indicate that the residues responsible for the functional diversification of type III PKSs are mutated in benzoic acid-specific type III PKSs. In order to gain a better understanding of structure-function relationships within the type III PKS family, the crystal structures of biphenyl synthase from Malus × domestica and benzophenone synthase from Hypericum androsaemum were compared with the structure of an archetypal type III PKS: chalcone synthase from Malus × domestica. Both biphenyl synthase and benzophenone synthase contain mutations that reshape their active-site cavities to prevent the binding of 4-coumaroyl-CoA and to favor the binding of small hydrophobic substrates. The active-site cavities of biphenyl synthase and benzophenone synthase also contain a novel pocket associated with their chain-elongation and cyclization reactions. Collectively, these results illuminate structural determinants of benzoic acid-specific type III PKSs and expand the understanding of the evolution of specialized metabolic pathways in plants.
Organizational Affiliation:
Howard Hughes Medical Institute, The Salk Institute for Biological Studies, La Jolla, CA 92037, USA.