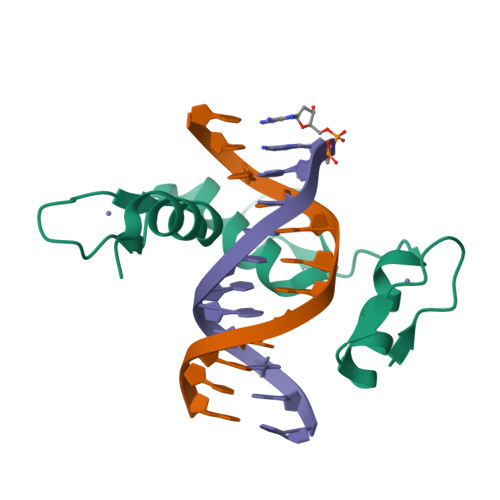
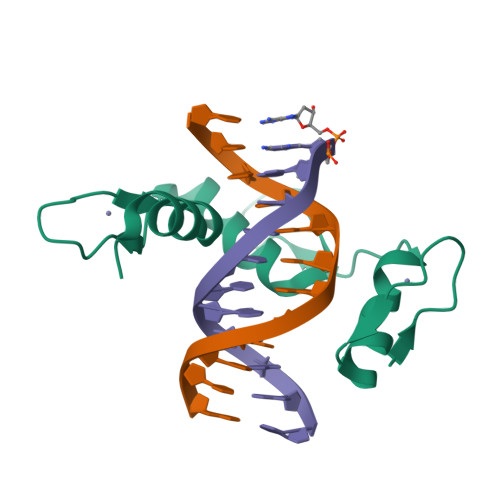
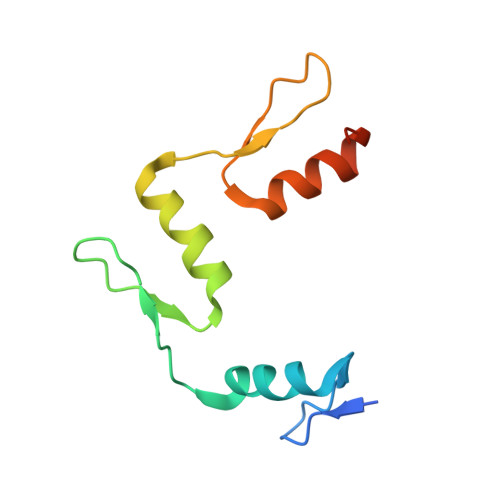
DNA methylation repels targeting of Arabidopsis REF6.
Qiu, Q., Mei, H., Deng, X., He, K., Wu, B., Yao, Q., Zhang, J., Lu, F., Ma, J., Cao, X.(2019) Nat Commun 10: 2063-2063
- PubMed: 31048693
- DOI: https://doi.org/10.1038/s41467-019-10026-1
- Primary Citation of Related Structures:
6JNL, 6JNM, 6JNN - PubMed Abstract:
RELATIVE OF EARLY FLOWERING 6 (REF6/JMJ12), a Jumonji C (JmjC)-domain-containing H3K27me3 histone demethylase, finds its target loci in Arabidopsis genome by directly recognizing the CTCTGYTY motif via its zinc-finger (ZnF) domains. REF6 tends to bind motifs located in active chromatin states that are depleted for heterochromatic modifications. However, the underlying mechanism remains unknown. Here, we show that REF6 preferentially bind to hypo-methylated CTCTGYTY motifs in vivo, and that CHG methylation decreases REF6 DNA binding affinity in vitro. In addition, crystal structures of ZnF-clusters in complex with DNA oligonucleotides reveal that 5-methylcytosine is unfavorable for REF6 binding. In drm1 drm2 cmt2 cmt3 (ddcc) quadruple mutants, in which non-CG methylation is significantly reduced, REF6 can ectopically bind a small number of new target loci, most of which are located in or neighbored with short TEs in euchromatic regions. Collectively, our findings reveal that DNA methylation, likely acting in combination with other epigenetic modifications, may partially explain why REF6 binding is depleted in heterochromatic loci.
Organizational Affiliation:
State Key Laboratory of Plant Genomics and National Center for Plant Gene Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, 100101, China.