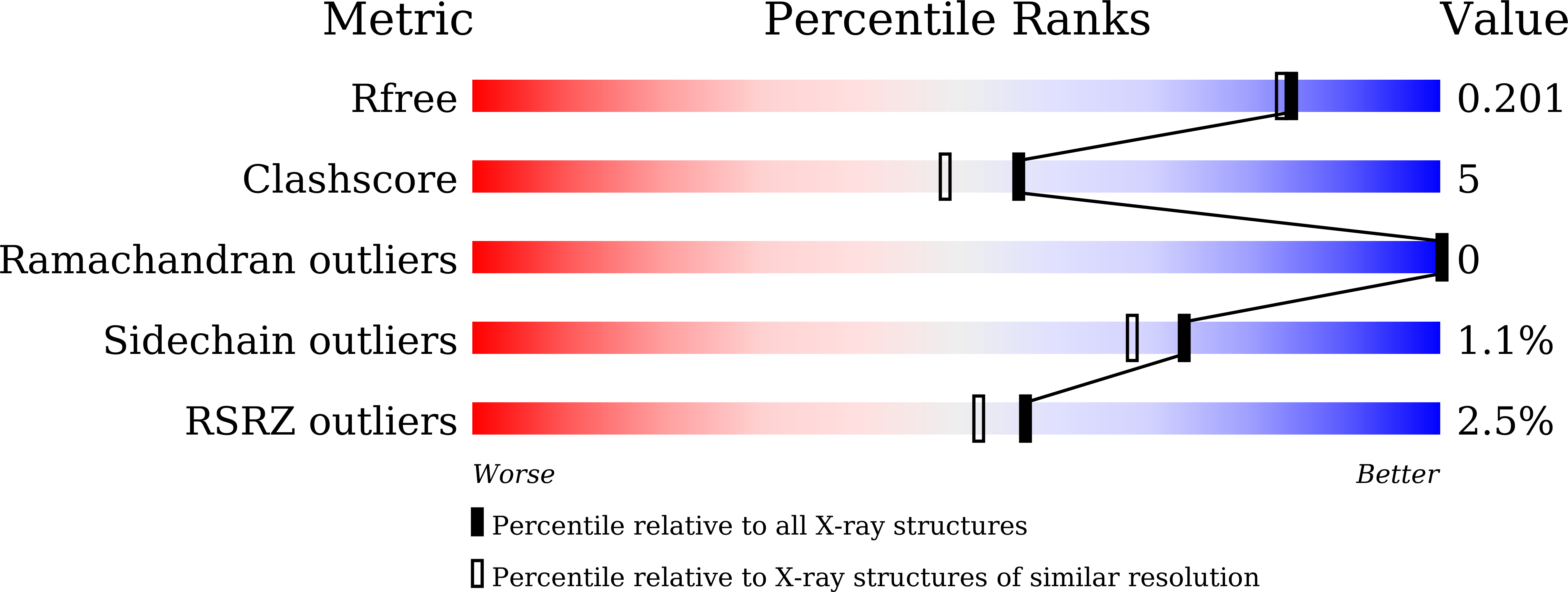
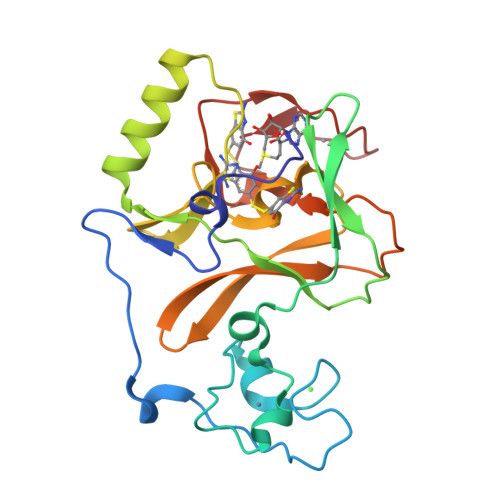
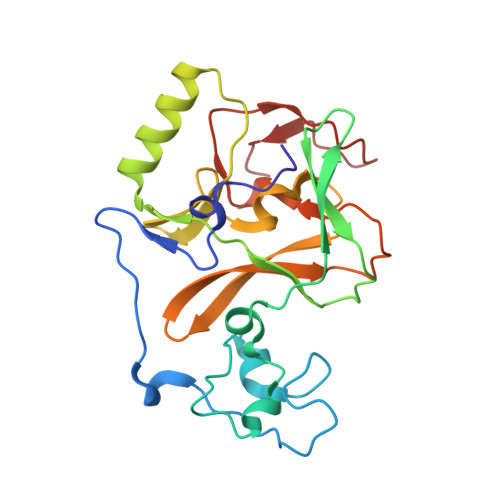
Covalent inhibition of NSD1 histone methyltransferase.
Huang, H., Howard, C.A., Zari, S., Cho, H.J., Shukla, S., Li, H., Ndoj, J., Gonzalez-Alonso, P., Nikolaidis, C., Abbott, J., Rogawski, D.S., Potopnyk, M.A., Kempinska, K., Miao, H., Purohit, T., Henderson, A., Mapp, A., Sulis, M.L., Ferrando, A., Grembecka, J., Cierpicki, T.(2020) Nat Chem Biol 16: 1403-1410
- PubMed: 32868895
- DOI: https://doi.org/10.1038/s41589-020-0626-6
- Primary Citation of Related Structures:
6KQP, 6KQQ - PubMed Abstract:
The nuclear receptor-binding SET domain (NSD) family of histone methyltransferases is associated with various malignancies, including aggressive acute leukemia with NUP98-NSD1 translocation. While NSD proteins represent attractive drug targets, their catalytic SET domains exist in autoinhibited conformation, presenting notable challenges for inhibitor development. Here, we employed a fragment-based screening strategy followed by chemical optimization, which resulted in the development of the first-in-class irreversible small-molecule inhibitors of the nuclear receptor-binding SET domain protein 1 (NSD1) SET domain. The crystal structure of NSD1 in complex with covalently bound ligand reveals a conformational change in the autoinhibitory loop of the SET domain and formation of a channel-like pocket suitable for targeting with small molecules. Our covalent lead-compound BT5-demonstrates on-target activity in NUP98-NSD1 leukemia cells, including inhibition of histone H3 lysine 36 dimethylation and downregulation of target genes, and impaired colony formation in an NUP98-NSD1 patient sample. This study will facilitate the development of the next generation of potent and selective inhibitors of the NSD histone methyltransferases.
Organizational Affiliation:
Department of Pathology, University of Michigan, Ann Arbor, MI, USA.