Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
Yao, Y., Huo, T., Lin, Y.L., Nie, S., Wu, F., Hua, Y., Wu, J., Kneubehl, A.R., Vogt, M.B., Rico-Hesse, R., Song, Y.(2019) J Am Chem Soc 141: 6832-6836
- PubMed: 31017399
- DOI: https://doi.org/10.1021/jacs.9b02505
- Primary Citation of Related Structures:
6MO0, 6MO1, 6MO2 - PubMed Abstract:
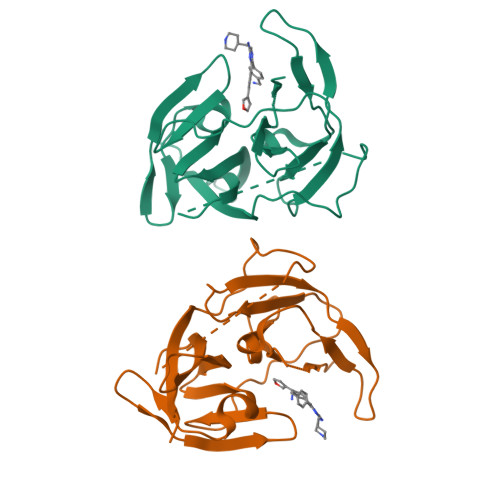
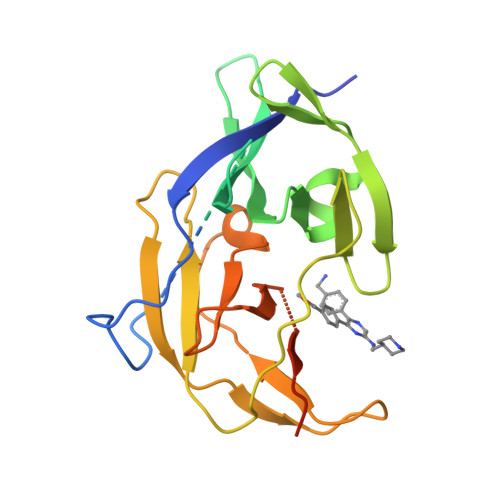
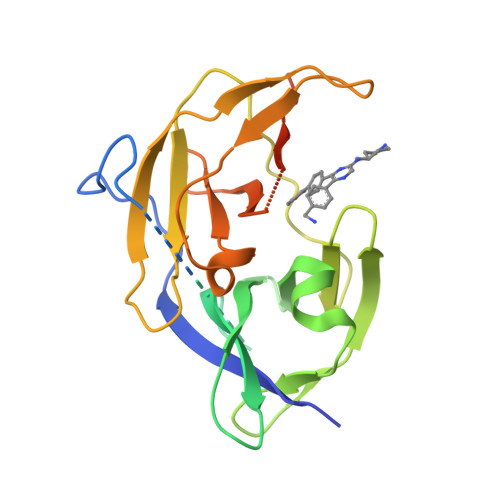
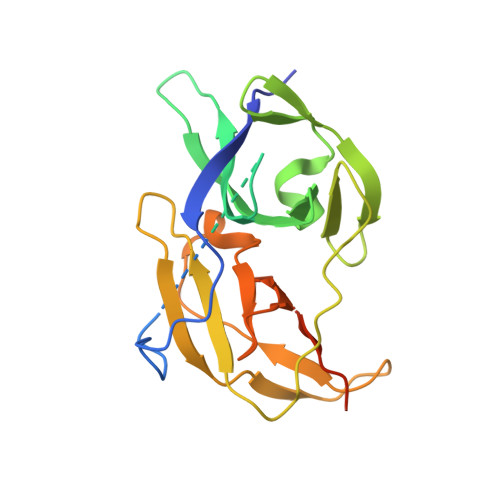
Flaviviruses, including dengue, West Nile and recently emerged Zika virus, are important human pathogens, but there are no drugs to prevent or treat these viral infections. The highly conserved Flavivirus NS2B-NS3 protease is essential for viral replication and therefore a drug target. Compound screening followed by medicinal chemistry yielded a series of drug-like, broadly active inhibitors of Flavivirus proteases with IC 50 as low as 120 nM. The inhibitor exhibited significant antiviral activities in cells (EC 68 : 300-600 nM) and in a mouse model of Zika virus infection. X-ray studies reveal that the inhibitors bind to an allosteric, mostly hydrophobic pocket of dengue NS3 and hold the protease in an open, catalytically inactive conformation. The inhibitors and their binding structures would be useful for rational drug development targeting Zika, dengue and other Flaviviruses.