Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Mashalidis, E.H., Kaeser, B., Terasawa, Y., Katsuyama, A., Kwon, D.Y., Lee, K., Hong, J., Ichikawa, S., Lee, S.Y.(2019) Nat Commun 10: 2917-2917
- PubMed: 31266949
- DOI: https://doi.org/10.1038/s41467-019-10957-9
- Primary Citation of Related Structures:
6OYH, 6OYZ, 6OZ6 - PubMed Abstract:
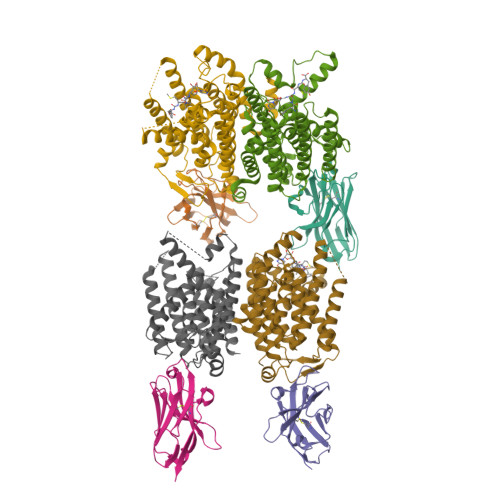
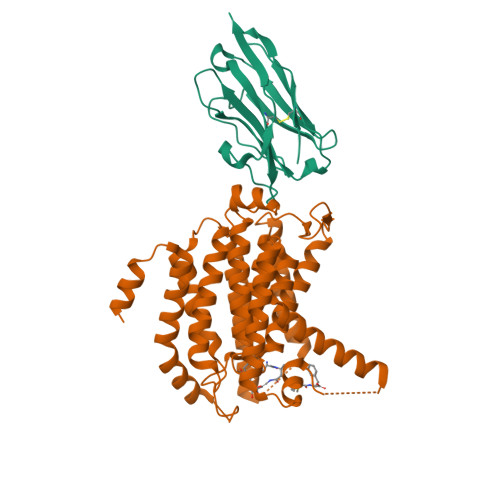
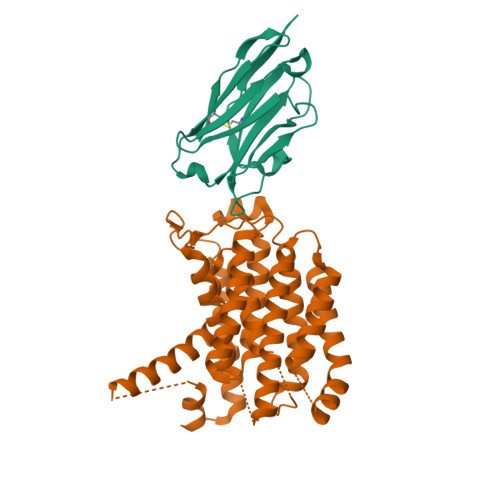
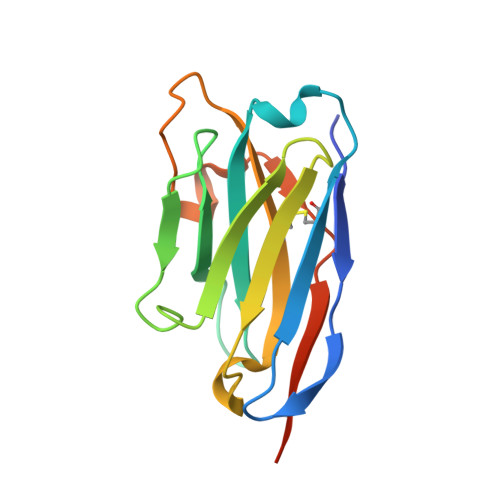
Novel antibacterial agents are needed to address the emergence of global antibiotic resistance. MraY is a promising candidate for antibiotic development because it is the target of five classes of naturally occurring nucleoside inhibitors with potent antibacterial activity. Although these natural products share a common uridine moiety, their core structures vary substantially and they exhibit different activity profiles. An incomplete understanding of the structural and mechanistic basis of MraY inhibition has hindered the translation of these compounds to the clinic. Here we present crystal structures of MraY in complex with representative members of the liposidomycin/caprazamycin, capuramycin, and mureidomycin classes of nucleoside inhibitors. Our structures reveal cryptic druggable hot spots in the shallow inhibitor binding site of MraY that were not previously appreciated. Structural analyses of nucleoside inhibitor binding provide insights into the chemical logic of MraY inhibition, which can guide novel approaches to MraY-targeted antibiotic design.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, 303 Research Drive, Durham, NC, 27710, USA.