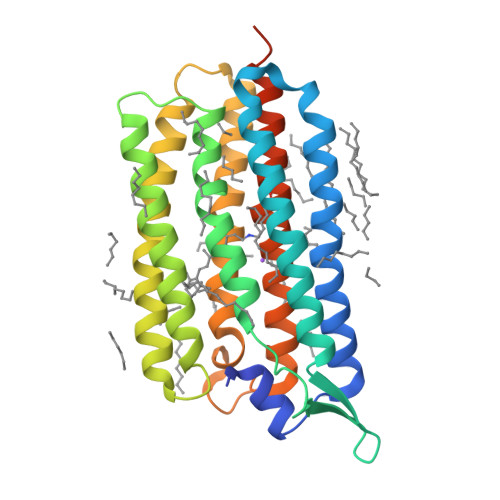
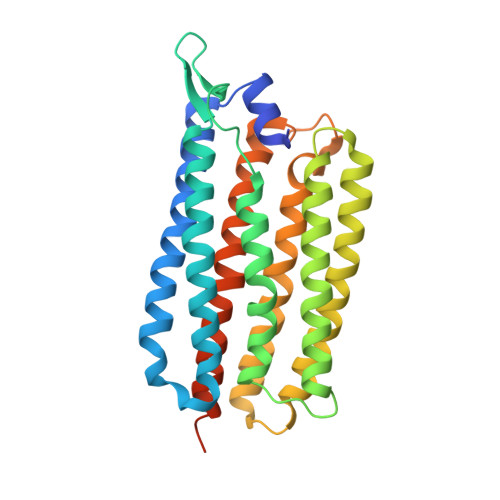
Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Skopintsev, P., Ehrenberg, D., Weinert, T., James, D., Kar, R.K., Johnson, P.J.M., Ozerov, D., Furrer, A., Martiel, I., Dworkowski, F., Nass, K., Knopp, G., Cirelli, C., Arrell, C., Gashi, D., Mous, S., Wranik, M., Gruhl, T., Kekilli, D., Brunle, S., Deupi, X., Schertler, G.F.X., Benoit, R.M., Panneels, V., Nogly, P., Schapiro, I., Milne, C., Heberle, J., Standfuss, J.(2020) Nature 583: 314-318
- PubMed: 32499654
- DOI: https://doi.org/10.1038/s41586-020-2307-8
- Primary Citation of Related Structures:
6TK1, 6TK2, 6TK3, 6TK4, 6TK5, 6TK6, 6TK7 - PubMed Abstract:
Light-driven sodium pumps actively transport small cations across cellular membranes 1 . These pumps are used by microorganisms to convert light into membrane potential and have become useful optogenetic tools with applications in neuroscience. Although the resting state structures of the prototypical sodium pump Krokinobacter eikastus rhodopsin 2 (KR2) have been solved 2,3 , it is unclear how structural alterations over time allow sodium to be translocated against a concentration gradient. Here, using the Swiss X-ray Free Electron Laser 4 , we have collected serial crystallographic data at ten pump-probe delays from femtoseconds to milliseconds. High-resolution structural snapshots throughout the KR2 photocycle show how retinal isomerization is completed on the femtosecond timescale and changes the local structure of the binding pocket in the early nanoseconds. Subsequent rearrangements and deprotonation of the retinal Schiff base open an electrostatic gate in microseconds. Structural and spectroscopic data, in combination with quantum chemical calculations, indicate that a sodium ion binds transiently close to the retinal within one millisecond. In the last structural intermediate, at 20 milliseconds after activation, we identified a potential second sodium-binding site close to the extracellular exit. These results provide direct molecular insight into the dynamics of active cation transport across biological membranes.
Organizational Affiliation:
Laboratory of Biomolecular Research, Biology and Chemistry Division, Paul Scherrer Institut, Villigen, Switzerland.