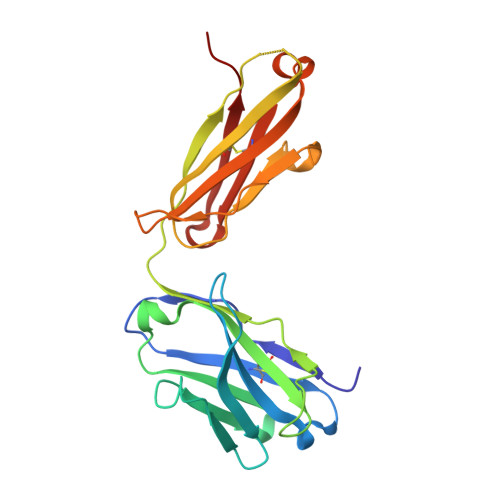
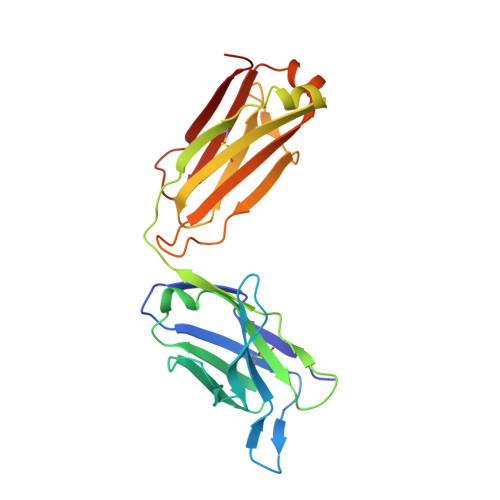
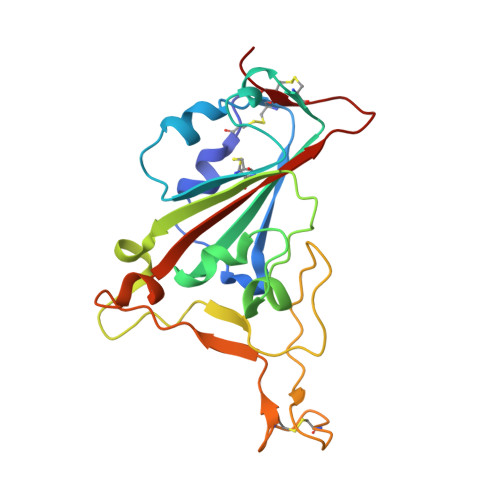
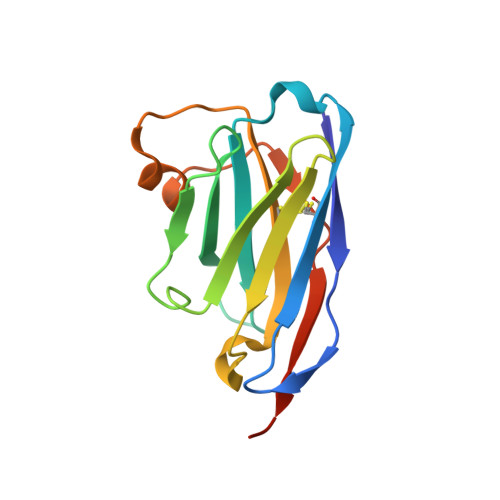
Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Huo, J., Le Bas, A., Ruza, R.R., Duyvesteyn, H.M.E., Mikolajek, H., Malinauskas, T., Tan, T.K., Rijal, P., Dumoux, M., Ward, P.N., Ren, J., Zhou, D., Harrison, P.J., Weckener, M., Clare, D.K., Vogirala, V.K., Radecke, J., Moynie, L., Zhao, Y., Gilbert-Jaramillo, J., Knight, M.L., Tree, J.A., Buttigieg, K.R., Coombes, N., Elmore, M.J., Carroll, M.W., Carrique, L., Shah, P.N.M., James, W., Townsend, A.R., Stuart, D.I., Owens, R.J., Naismith, J.H.(2020) Nat Struct Mol Biol 27: 846-854
- PubMed: 32661423
- DOI: https://doi.org/10.1038/s41594-020-0469-6
- Primary Citation of Related Structures:
6ZH9 - PubMed Abstract:
The SARS-CoV-2 virus is more transmissible than previous coronaviruses and causes a more serious illness than influenza. The SARS-CoV-2 receptor binding domain (RBD) of the spike protein binds to the human angiotensin-converting enzyme 2 (ACE2) receptor as a prelude to viral entry into the cell. Using a naive llama single-domain antibody library and PCR-based maturation, we have produced two closely related nanobodies, H11-D4 and H11-H4, that bind RBD (K D of 39 and 12 nM, respectively) and block its interaction with ACE2. Single-particle cryo-EM revealed that both nanobodies bind to all three RBDs in the spike trimer. Crystal structures of each nanobody-RBD complex revealed how both nanobodies recognize the same epitope, which partly overlaps with the ACE2 binding surface, explaining the blocking of the RBD-ACE2 interaction. Nanobody-Fc fusions showed neutralizing activity against SARS-CoV-2 (4-6 nM for H11-H4, 18 nM for H11-D4) and additive neutralization with the SARS-CoV-1/2 antibody CR3022.
Organizational Affiliation:
Structural Biology, The Rosalind Franklin Institute, Harwell Science & Innovation Campus, Didcot, UK.