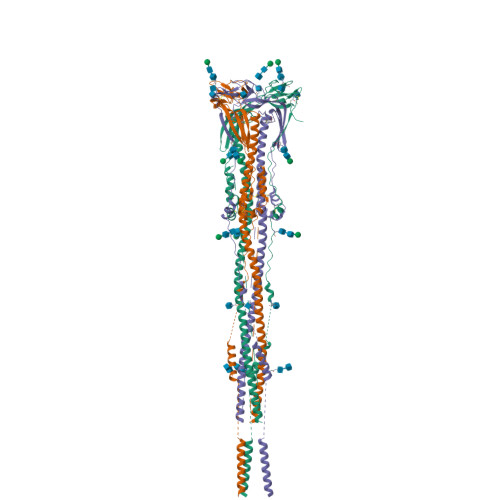
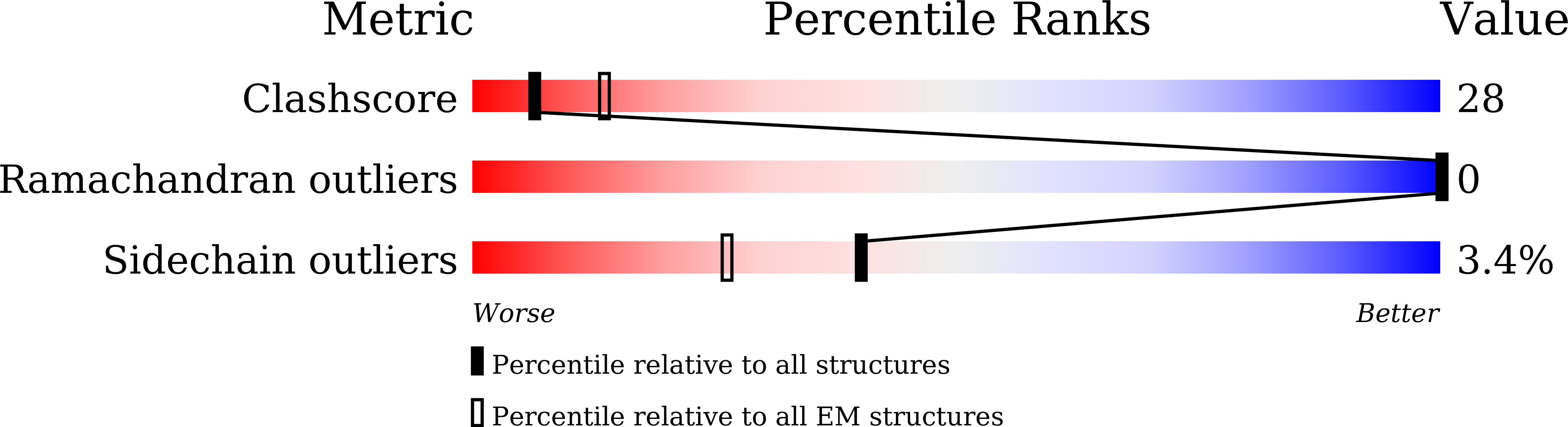Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein.
Tai, L., Zhu, G., Yang, M., Cao, L., Xing, X., Yin, G., Chan, C., Qin, C., Rao, Z., Wang, X., Sun, F., Zhu, Y.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34782481
- DOI: https://doi.org/10.1073/pnas.2112703118
- Primary Citation of Related Structures:
7E9T - PubMed Abstract:
The spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mediates membrane fusion to allow entry of the viral genome into host cells. To understand its detailed entry mechanism and develop a specific entry inhibitor, in situ structural information on the SARS-CoV-2 spike protein in different states is urgent. Here, by using cryo-electron tomography, we observed both prefusion and postfusion spikes in β-propiolactone-inactivated SARS-CoV-2 virions and solved the in situ structure of the postfusion spike at nanometer resolution. Compared to previous reports, the six-helix bundle fusion core, the glycosylation sites, and the location of the transmembrane domain were clearly resolved. We observed oligomerization patterns of the spikes on the viral membrane, likely suggesting a mechanism of fusion pore formation.
Organizational Affiliation:
National Key Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.