RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Liang, Q., Wang, Y., Zhang, S., Sun, J., Sun, W., Li, J., Liu, Y., Li, M., Cheng, L., Jiang, Y., Wang, R., Zhang, R., Yang, Z., Ren, Y., Chen, P., Gao, P., Yan, H., Zhang, Z., Zhang, Q., Shi, X., Wang, J., Liu, W., Wang, X., Ying, B., Zhao, J., Qi, H., Zhang, L.(2022) iScience 25: 104043-104043
- PubMed: 35291264
- DOI: https://doi.org/10.1016/j.isci.2022.104043
- Primary Citation of Related Structures:
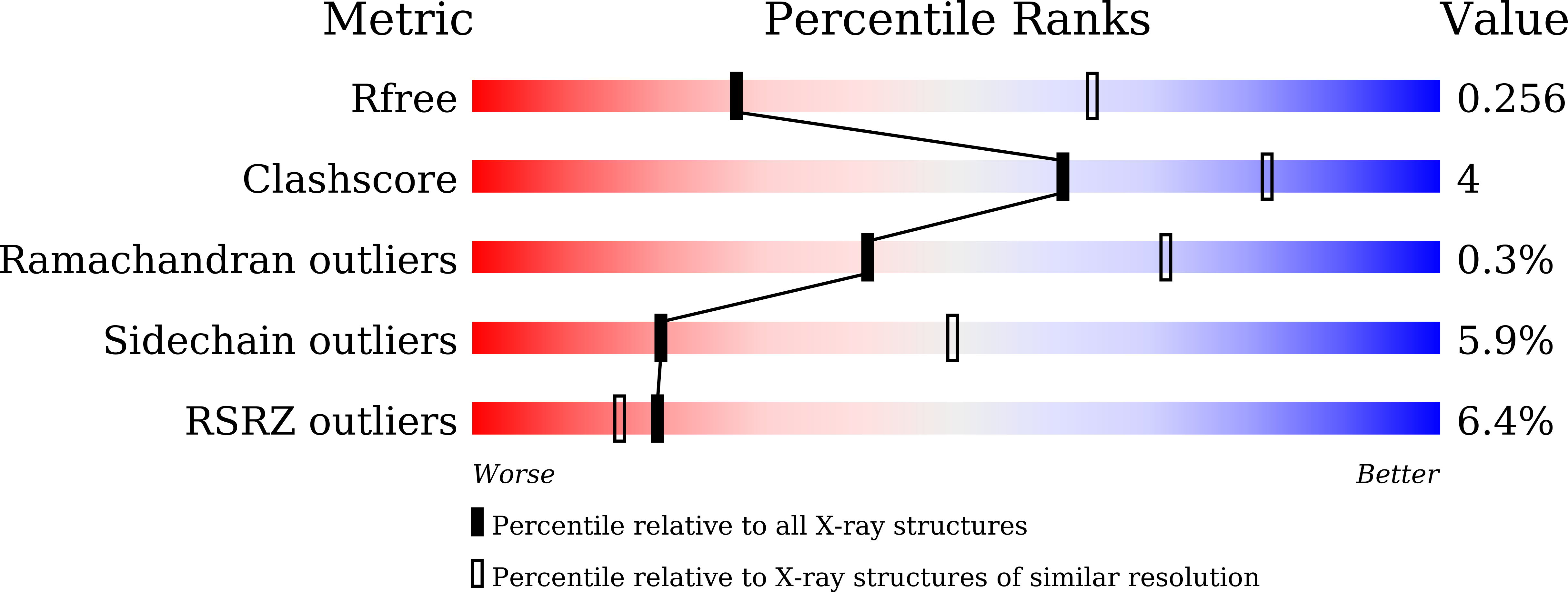
7FJN, 7FJO, 7FJS - PubMed Abstract:
With the rapid emergence and spread of SARS-CoV-2 variants, development of vaccines with broad and potent protectivity has become a global priority. Here, we designed a lipid nanoparticle-encapsulated, nucleoside-unmodified mRNA (mRNA-LNP) vaccine encoding the trimerized receptor-binding domain (RBD trimer) and showed its robust capability in inducing broad and protective immune responses against wild-type and major variants of concern (VOCs) in the mouse model of SARS-CoV-2 infection. The protectivity was correlated with RBD-specific B cell responses especially the long-lived plasma B cells in bone marrow, strong ability in triggering BCR clustering, and downstream signaling. Monoclonal antibodies isolated from vaccinated animals demonstrated broad and potent neutralizing activity against VOCs tested. Structure analysis of one representative antibody identified a novel epitope with a high degree of conservation among different variants. Collectively, these results demonstrate that the RBD trimer mRNA vaccine serves as a promising vaccine candidate against SARS-CoV-2 variants and beyond.
Organizational Affiliation:
Comprehensive AIDS Research Center, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.