Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP.
Biswal, M., Diggs, S., Xu, D., Khudaverdyan, N., Lu, J., Fang, J., Blaha, G., Hai, R., Song, J.(2021) Nucleic Acids Res 49: 5956-5966
- PubMed: 33999154
- DOI: https://doi.org/10.1093/nar/gkab370
- Primary Citation of Related Structures:
7JLT - PubMed Abstract:
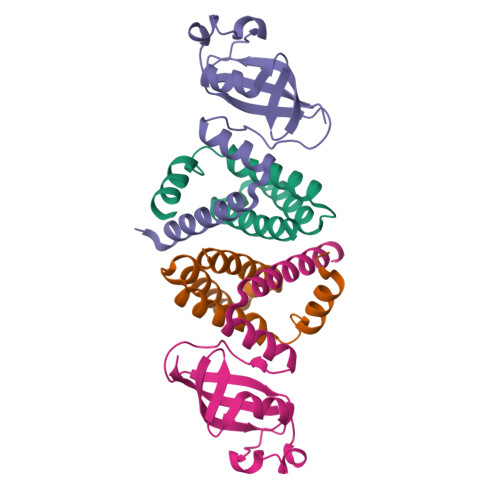
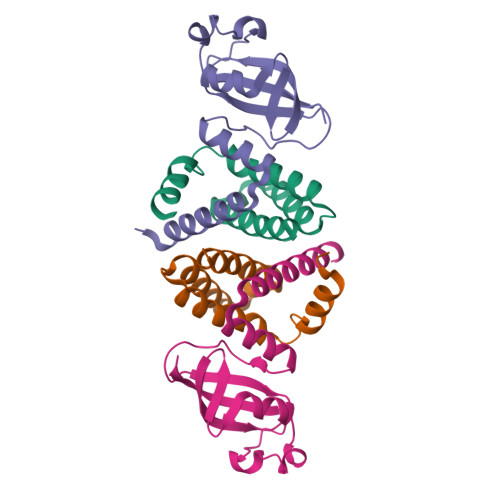
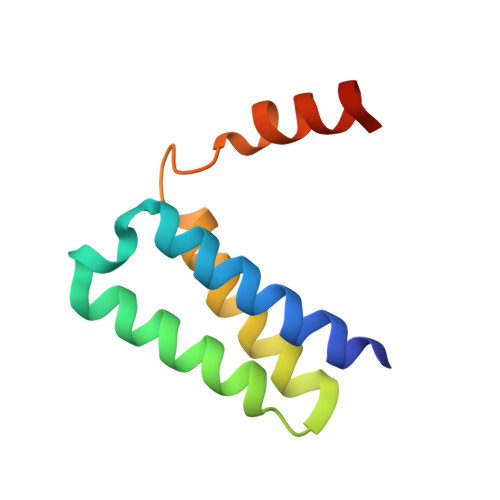
Replication of the ∼30 kb-long coronavirus genome is mediated by a complex of non-structural proteins (NSP), in which NSP7 and NSP8 play a critical role in regulating the RNA-dependent RNA polymerase (RdRP) activity of NSP12. The assembly of NSP7, NSP8 and NSP12 proteins is highly dynamic in solution, yet the underlying mechanism remains elusive. We report the crystal structure of the complex between NSP7 and NSP8 of SARS-CoV-2, revealing a 2:2 heterotetrameric form. Formation of the NSP7-NSP8 complex is mediated by two distinct oligomer interfaces, with interface I responsible for heterodimeric NSP7-NSP8 assembly, and interface II mediating the heterotetrameric interaction between the two NSP7-NSP8 dimers. Structure-guided mutagenesis, combined with biochemical and enzymatic assays, further reveals a structural coupling between the two oligomer interfaces, as well as the importance of these interfaces for the RdRP activity of the NSP7-NSP8-NSP12 complex. Finally, we identify an NSP7 mutation that differentially affects the stability of the NSP7-NSP8 and NSP7-NSP8-NSP12 complexes leading to a selective impairment of the RdRP activity. Together, this study provides deep insights into the structure and mechanism for the dynamic assembly of NSP7 and NSP8 in regulating the replication of the SARS-CoV-2 genome, with important implications for antiviral drug development.
Organizational Affiliation:
Department of Biochemistry, University of California-Riverside, Riverside, CA, USA.