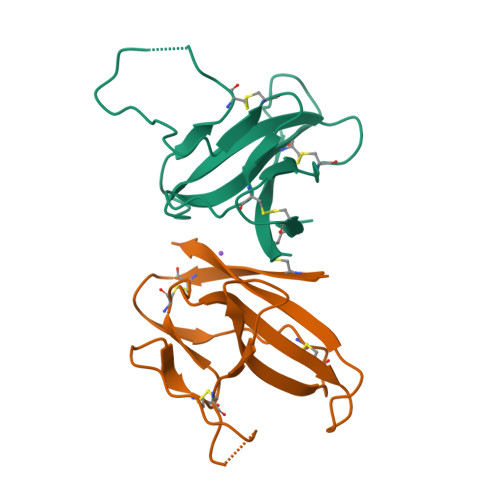
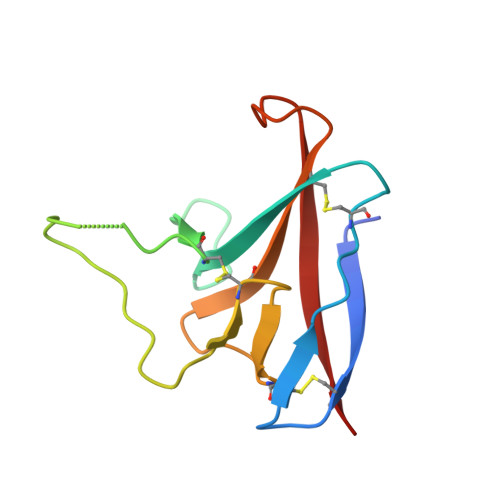
Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein.
Flower, T.G., Buffalo, C.Z., Hooy, R.M., Allaire, M., Ren, X., Hurley, J.H.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33361333
- DOI: https://doi.org/10.1073/pnas.2021785118
- Primary Citation of Related Structures:
7JTL - PubMed Abstract:
The molecular basis for the severity and rapid spread of the COVID-19 disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is largely unknown. ORF8 is a rapidly evolving accessory protein that has been proposed to interfere with immune responses. The crystal structure of SARS-CoV-2 ORF8 was determined at 2.04-Å resolution by X-ray crystallography. The structure reveals a ∼60-residue core similar to SARS-CoV-2 ORF7a, with the addition of two dimerization interfaces unique to SARS-CoV-2 ORF8. A covalent disulfide-linked dimer is formed through an N-terminal sequence specific to SARS-CoV-2, while a separate noncovalent interface is formed by another SARS-CoV-2-specific sequence, 73 YIDI 76 Together, the presence of these interfaces shows how SARS-CoV-2 ORF8 can form unique large-scale assemblies not possible for SARS-CoV, potentially mediating unique immune suppression and evasion activities.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720.