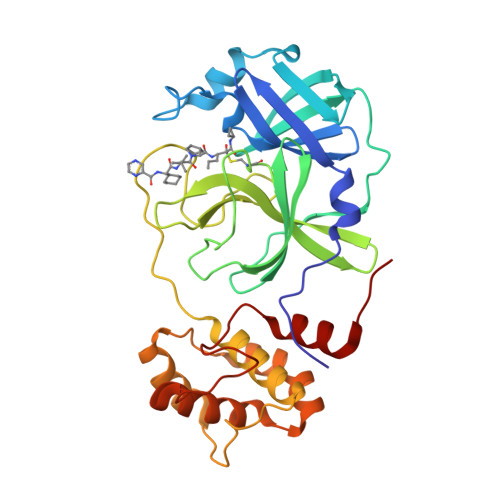
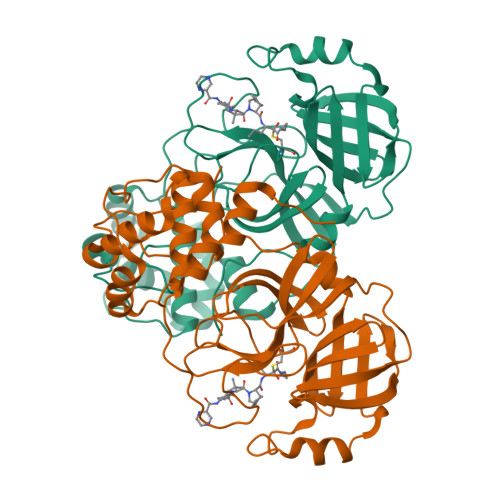
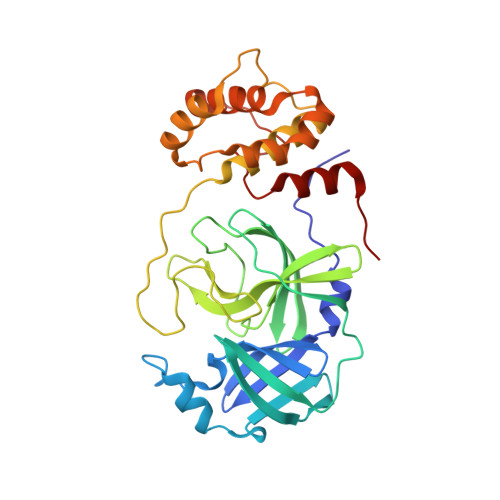
Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
Kneller, D.W., Phillips, G., Weiss, K.L., Zhang, Q., Coates, L., Kovalevsky, A.(2021) J Med Chem 64: 4991-5000
- PubMed: 33755450
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00058
- Primary Citation of Related Structures:
7LB7 - PubMed Abstract:
The main protease (3CL M pro ) from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, is an essential enzyme for viral replication with no human counterpart, making it an attractive drug target. To date, no small-molecule clinical drugs are available that specifically inhibit SARS-CoV-2 M pro . To aid rational drug design, we determined a neutron structure of M pro in complex with the α-ketoamide inhibitor telaprevir at near-physiological (22 °C) temperature. We directly observed protonation states in the inhibitor complex and compared them with those in the ligand-free M pro , revealing modulation of the active-site protonation states upon telaprevir binding. We suggest that binding of other α-ketoamide covalent inhibitors can lead to the same protonation state changes in the M pro active site. Thus, by studying the protonation state changes induced by inhibitors, we provide crucial insights to help guide rational drug design, allowing precise tailoring of inhibitors to manipulate the electrostatic environment of SARS-CoV-2 M pro .
Organizational Affiliation:
Neutron Scattering Division, Oak Ridge National Laboratory, 1 Bethel Valley Road, Oak Ridge, Tennessee 37831, United States.