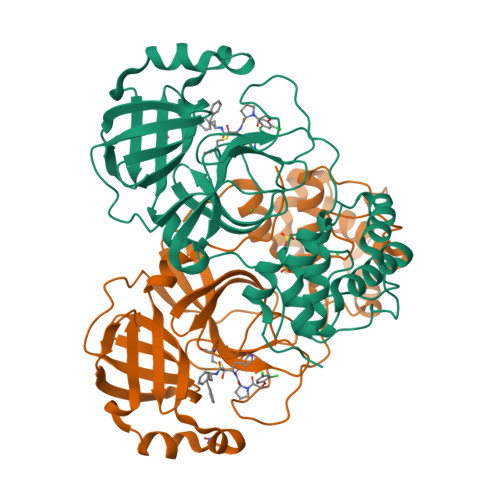
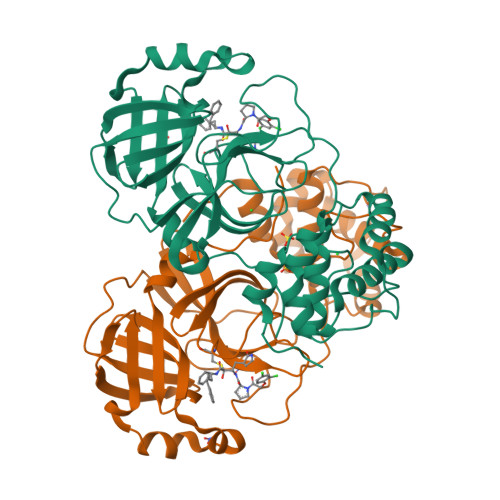
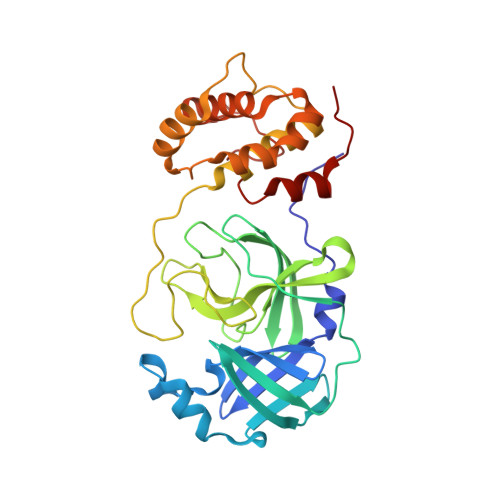
Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Pelliccia, S., Cerchia, C., Esposito, F., Cannalire, R., Corona, A., Costanzi, E., Kuzikov, M., Gribbon, P., Zaliani, A., Brindisi, M., Storici, P., Tramontano, E., Summa, V.(2022) Eur J Med Chem 244: 114853-114853
- PubMed: 36332546
- DOI: https://doi.org/10.1016/j.ejmech.2022.114853
- Primary Citation of Related Structures:
7Q5E, 7Q5F - PubMed Abstract:
SARS-CoV-2 caused worldwide the current outbreak called COVID-19. Despite multiple countermeasures implemented, there is an urgent global need for new potent and efficient antiviral drugs against this pathogen. In this context, the main protease (M pro ) of SARS-CoV-2 is an essential viral enzyme and plays a pivotal role in viral replication and transcription. Its specific cleavage of polypeptides after a glutamine residue has been considered as a key element to design novel antiviral drugs. Herein, we reported the design, synthesis and structure-activity relationships of novel α-ketoamides as covalent reversible inhibitors of M pro , exploiting the PADAM oxidation route. The reported compounds showed μM to nM activities in enzymatic and in the antiviral cell-based assays against SARS-CoV-2 M pro . In order to assess inhibitors' binding mode, two co-crystal structures of SARS-CoV-2 M pro in complex with our inhibitors were solved, which confirmed the covalent binding of the keto amide moiety to the catalytic Cys145 residue of M pro . Finally, in order to interrogate potential broad-spectrum properties, we assessed a selection of compounds against MERS M pro where they showed nM inhibitory potency, thus highlighting their potential as broad-spectrum coronavirus inhibitors.
Organizational Affiliation:
Department of Pharmacy, University of Naples Federico II, via D. Montesano 49, 80131, Naples, Italy. Electronic address: sveva.pelliccia@unina.it.