[1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
Murthy, S., Nizi, M.G., Maksimainen, M.M., Massari, S., Alaviuhkola, J., Lippok, B.E., Vagaggini, C., Sowa, S.T., Galera-Prat, A., Ashok, Y., Venkannagari, H., Prunskaite-Hyyrylainen, R., Dreassi, E., Luscher, B., Korn, P., Tabarrini, O., Lehtio, L.(2023) J Med Chem 66: 1301-1320
- PubMed: 36598465
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01460
- Primary Citation of Related Structures:
7R3L, 7R3O, 7R3Z, 7R4A, 7R59, 7R5D, 7R5X, 7Z1V, 7Z1W, 7Z1Y, 7Z2O, 7Z2Q, 7Z41 - PubMed Abstract:
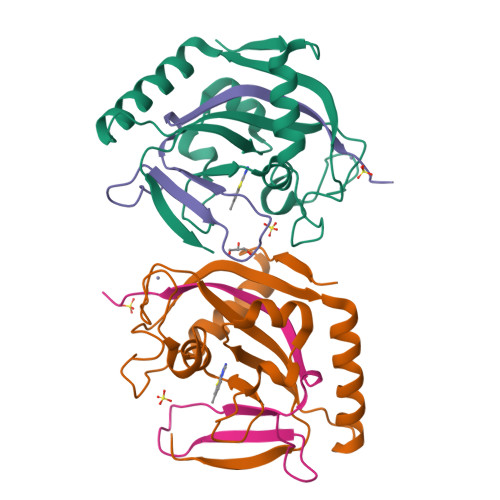
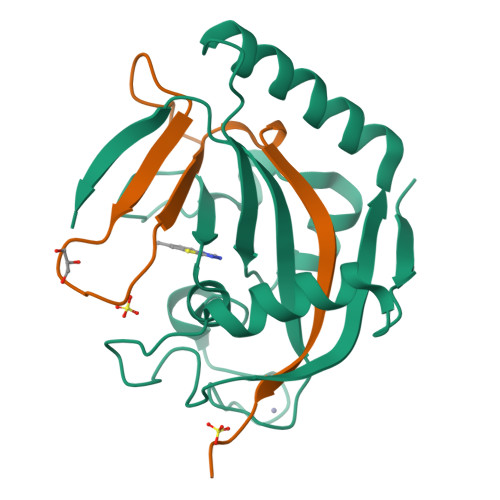
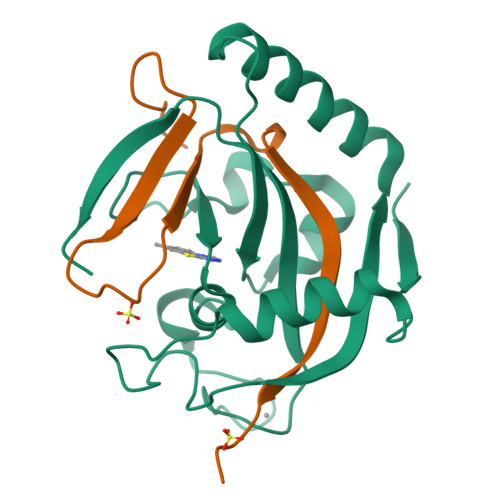
We report [1,2,4]triazolo[3,4- b ]benzothiazole (TBT) as a new inhibitor scaffold, which competes with nicotinamide in the binding pocket of human poly- and mono-ADP-ribosylating enzymes. The binding mode was studied through analogues and cocrystal structures with TNKS2, PARP2, PARP14, and PARP15. Based on the substitution pattern, we were able to identify 3-amino derivatives 21 (OUL243) and 27 (OUL232) as inhibitors of mono-ARTs PARP7, PARP10, PARP11, PARP12, PARP14, and PARP15 at nM potencies, with 27 being the most potent PARP10 inhibitor described to date (IC 50 of 7.8 nM) and the first PARP12 inhibitor ever reported. On the contrary, hydroxy derivative 16 (OUL245) inhibits poly-ARTs with a selectivity toward PARP2. The scaffold does not possess inherent cell toxicity, and the inhibitors can enter cells and engage with the target protein. This, together with favorable ADME properties, demonstrates the potential of TBT scaffold for future drug development efforts toward selective inhibitors against specific enzymes.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine and Biocenter Oulu, University of Oulu, Oulu90220, Finland.