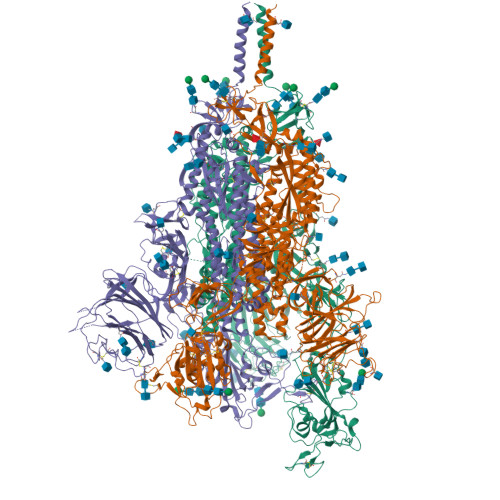
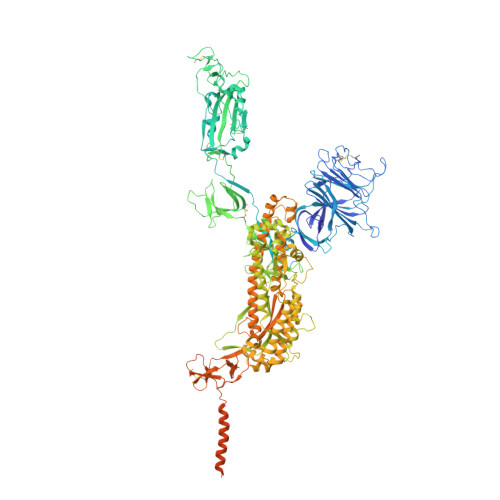
Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Zhang, J., Xiao, T., Cai, Y., Lavine, C.L., Peng, H., Zhu, H., Anand, K., Tong, P., Gautam, A., Mayer, M.L., Walsh Jr., R.M., Rits-Volloch, S., Wesemann, D.R., Yang, W., Seaman, M.S., Lu, J., Chen, B.(2021) Science 374: 1353-1360
- PubMed: 34698504
- DOI: https://doi.org/10.1126/science.abl9463
- Primary Citation of Related Structures:
7SBK, 7SBL, 7SBO, 7SBP, 7SBQ, 7SBR, 7SBS, 7SBT - PubMed Abstract:
The Delta variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has outcompeted previously prevalent variants and become a dominant strain worldwide. We report the structure, function, and antigenicity of its full-length spike (S) trimer as well as those of the Gamma and Kappa variants, and compare their characteristics with the G614, Alpha, and Beta variants. Delta S can fuse membranes more efficiently at low levels of cellular receptor angiotensin converting enzyme 2 (ACE2), and its pseudotyped viruses infect target cells substantially faster than the other five variants, possibly accounting for its heightened transmissibility. Each variant shows different rearrangement of the antigenic surface of the amino-terminal domain of the S protein but only makes produces changes in the receptor binding domain (RBD), making the RBD a better target for therapeutic antibodies.
Organizational Affiliation:
Division of Molecular Medicine, Boston Children's Hospital, Harvard Medical School, 3 Blackfan Street, Boston, MA 02115, USA.