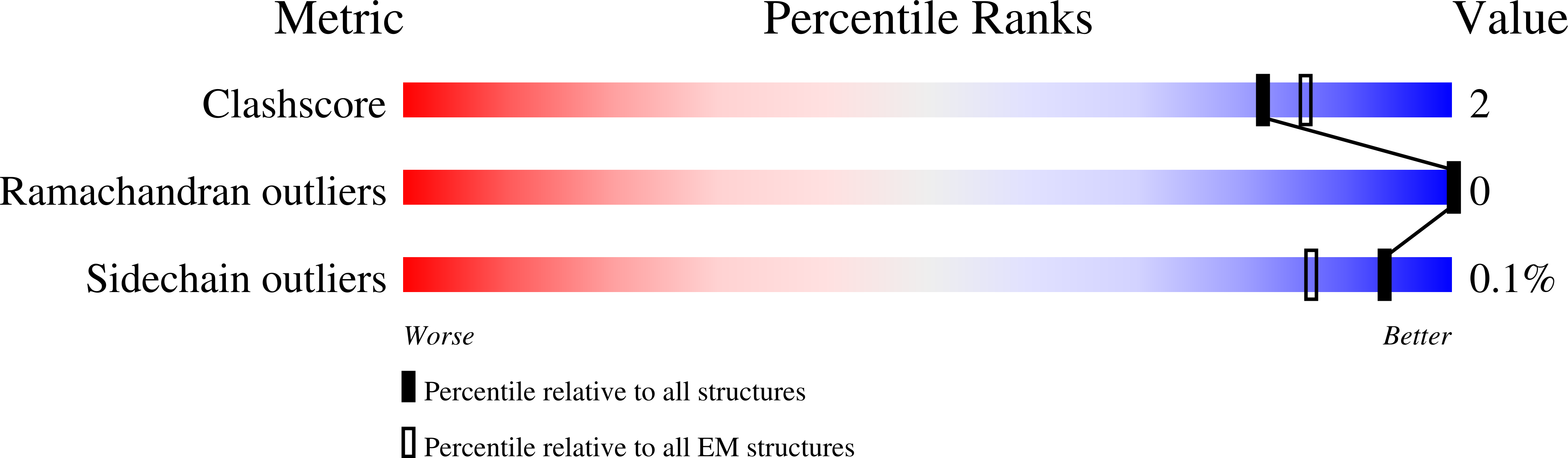Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Saville, J.W., Mannar, D., Zhu, X., Srivastava, S.S., Berezuk, A.M., Demers, J.P., Zhou, S., Tuttle, K.S., Sekirov, I., Kim, A., Li, W., Dimitrov, D.S., Subramaniam, S.(2022) Nat Commun 13: 742-742
- PubMed: 35136050
- DOI: https://doi.org/10.1038/s41467-022-28324-6
- Primary Citation of Related Structures:
7TEW, 7TEX, 7TEY, 7TEZ, 7TF0, 7TF1, 7TF2, 7TF3, 7TF4, 7TF5 - PubMed Abstract:
The Delta and Kappa variants of SARS-CoV-2 co-emerged in India in late 2020, with the Delta variant underlying the resurgence of COVID-19, even in countries with high vaccination rates. In this study, we assess structural and biochemical aspects of viral fitness for these two variants using cryo-electron microscopy (cryo-EM), ACE2-binding and antibody neutralization analyses. Both variants demonstrate escape of antibodies targeting the N-terminal domain, an important immune hotspot for neutralizing epitopes. Compared to wild-type and Kappa lineages, Delta variant spike proteins show modest increase in ACE2 affinity, likely due to enhanced electrostatic complementarity at the RBD-ACE2 interface, which we characterize by cryo-EM. Unexpectedly, Kappa variant spike trimers form a structural head-to-head dimer-of-trimers assembly, which we demonstrate is a result of the E484Q mutation and with unknown biological implications. The combination of increased antibody escape and enhanced ACE2 binding provides an explanation, in part, for the rapid global dominance of the Delta variant.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, V6T 1Z3, Canada.