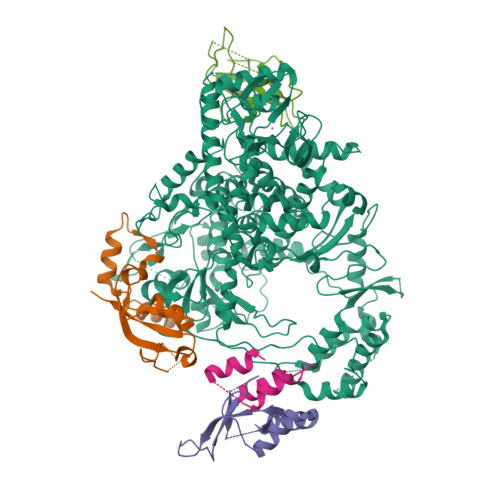
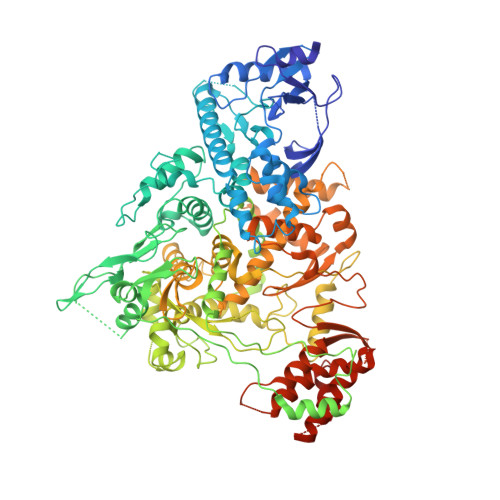
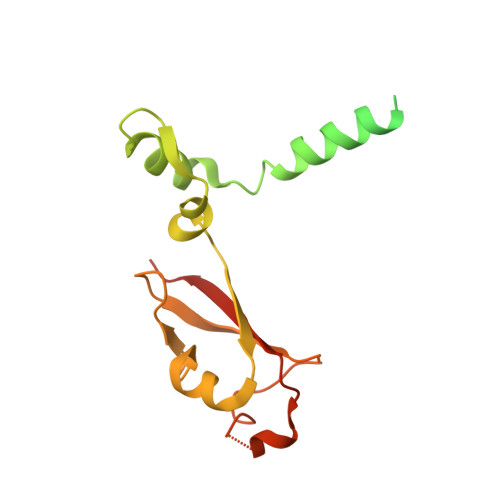
The mechanism of RNA capping by SARS-CoV-2.
Park, G.J., Osinski, A., Hernandez, G., Eitson, J.L., Majumdar, A., Tonelli, M., Henzler-Wildman, K., Pawlowski, K., Chen, Z., Li, Y., Schoggins, J.W., Tagliabracci, V.S.(2022) Nature 609: 793-800
- PubMed: 35944563
- DOI: https://doi.org/10.1038/s41586-022-05185-z
- Primary Citation of Related Structures:
7THM - PubMed Abstract:
The RNA genome of SARS-CoV-2 contains a 5' cap that facilitates the translation of viral proteins, protection from exonucleases and evasion of the host immune response 1-4 . How this cap is made in SARS-CoV-2 is not completely understood. Here we reconstitute the N7- and 2'-O-methylated SARS-CoV-2 RNA cap ( 7Me GpppA 2'-O-Me ) using virally encoded non-structural proteins (nsps). We show that the kinase-like nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain 5 of nsp12 transfers the RNA to the amino terminus of nsp9, forming a covalent RNA-protein intermediate (a process termed RNAylation). Subsequently, the NiRAN domain transfers the RNA to GDP, forming the core cap structure GpppA-RNA. The nsp14 6 and nsp16 7 methyltransferases then add methyl groups to form functional cap structures. Structural analyses of the replication-transcription complex bound to nsp9 identified key interactions that mediate the capping reaction. Furthermore, we demonstrate in a reverse genetics system 8 that the N terminus of nsp9 and the kinase-like active-site residues in the NiRAN domain are required for successful SARS-CoV-2 replication. Collectively, our results reveal an unconventional mechanism by which SARS-CoV-2 caps its RNA genome, thus exposing a new target in the development of antivirals to treat COVID-19.
Organizational Affiliation:
Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, USA.