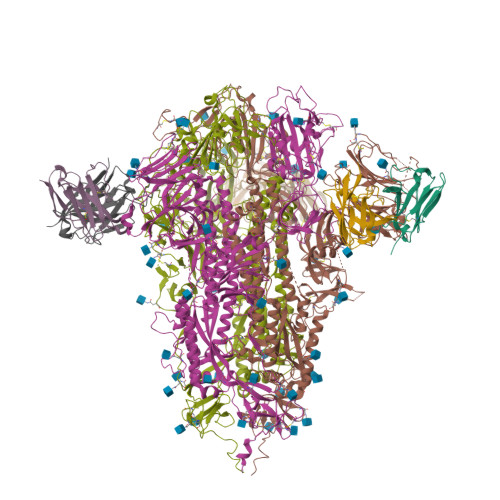
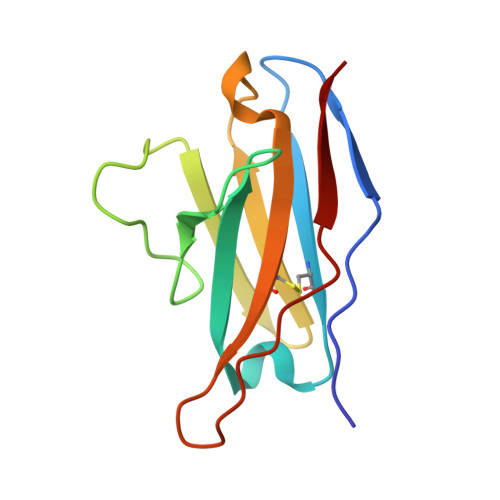
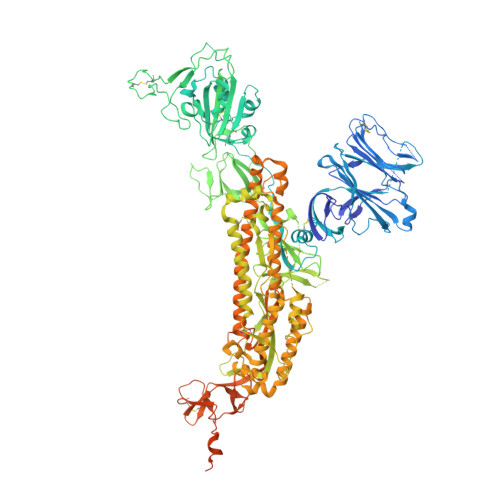
Antibodies targeting a quaternary site on SARS-CoV-2 spike glycoprotein prevent viral receptor engagement by conformational locking.
Liu, L., Casner, R.G., Guo, Y., Wang, Q., Iketani, S., Chan, J.F., Yu, J., Dadonaite, B., Nair, M.S., Mohri, H., Reddem, E.R., Yuan, S., Poon, V.K., Chan, C.C., Yuen, K.Y., Sheng, Z., Huang, Y., Bloom, J.D., Shapiro, L., Ho, D.D.(2023) Immunity 56: 2442
- PubMed: 37776849
- DOI: https://doi.org/10.1016/j.immuni.2023.09.003
- Primary Citation of Related Structures:
7UKL - PubMed Abstract:
SARS-CoV-2 continues to evolve, with many variants evading clinically authorized antibodies. To isolate monoclonal antibodies (mAbs) with broadly neutralizing capacities against the virus, we screened serum samples from convalescing COVID-19 patients. We isolated two mAbs, 12-16 and 12-19, which neutralized all SARS-CoV-2 variants tested, including the XBB subvariants, and prevented infection in hamsters challenged with Omicron BA.1 intranasally. Structurally, both antibodies targeted a conserved quaternary epitope located at the interface between the N-terminal domain and subdomain 1, uncovering a site of vulnerability on SARS-CoV-2 spike. These antibodies prevented viral receptor engagement by locking the receptor-binding domain (RBD) of spike in the down conformation, revealing a mechanism of virus neutralization for non-RBD antibodies. Deep mutational scanning showed that SARS-CoV-2 could mutate to escape 12-19, but such mutations are rarely found in circulating viruses. Antibodies 12-16 and 12-19 hold promise as prophylactic agents for immunocompromised persons who do not respond robustly to COVID-19 vaccines.
Organizational Affiliation:
Aaron Diamond AIDS Research Center, Columbia University Vagelos College of Physicians and Surgeons, New York, NY 10032, USA; Division of Infectious Diseases, Department of Medicine, Columbia University Vagelos College of Physicians and Surgeons, New York, NY 10032, USA. Electronic address: ll3411@cumc.columbia.edu.