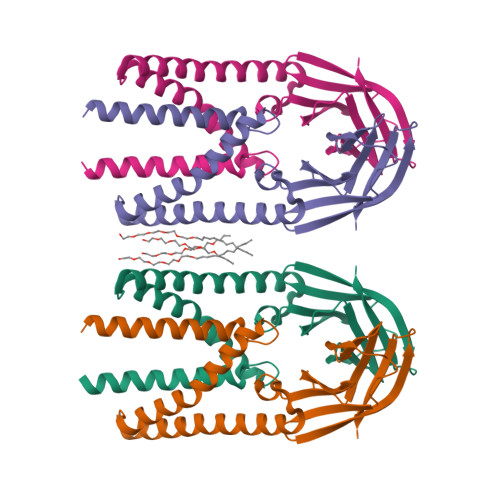
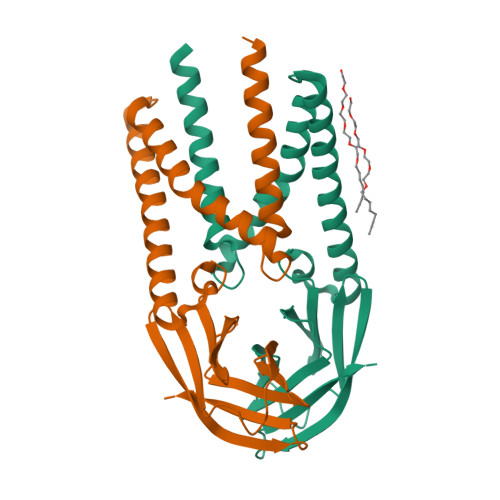
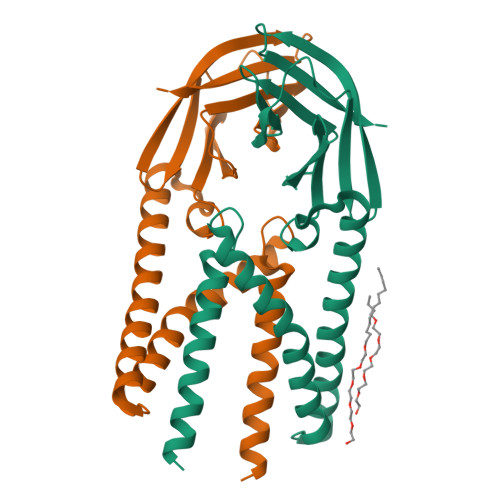
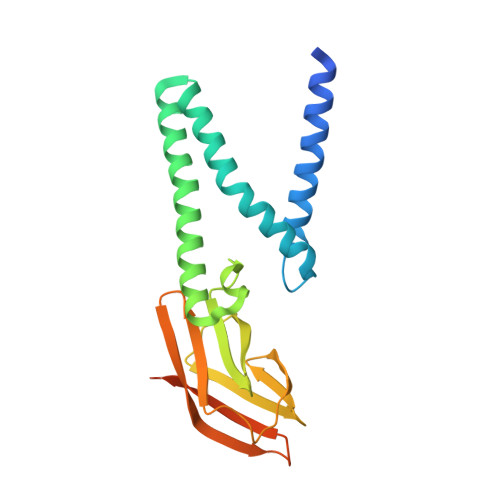
Crystal structure of the membrane (M) protein from a bat betacoronavirus.
Wang, X., Yang, Y., Sun, Z., Zhou, X.(2023) PNAS Nexus 2: pgad021-pgad021
- PubMed: 36874273
- DOI: https://doi.org/10.1093/pnasnexus/pgad021
- Primary Citation of Related Structures:
7Y96, 7Y9B - PubMed Abstract:
The membrane (M) protein is the most abundant structural protein of coronaviruses including MERS-CoV, SARS-CoV, and SARS-CoV-2, and plays a central role in virus assembly through its interaction with various partner proteins. However, mechanistic details about how M protein interacts with others remain elusive due to lack of high-resolution structures. Here, we present the first crystal structure of a betacoronavirus M protein from Pipistrellus bat coronavirus HKU5 (batCOV5-M), which is closely related to MERS-CoV, SARS-CoV, and SARS-CoV-2 M proteins. Furthermore, an interaction analysis indicates that the carboxy-terminus of the batCOV5 nucleocapsid (N) protein mediates its interaction with batCOV5-M. Combined with a computational docking analysis an M-N interaction model is proposed, providing insight into the mechanism of M protein-mediated protein interactions.
Organizational Affiliation:
State Key Laboratory of Biotherapy, Department of Integrated Traditional Chinese and Western Medicine, Rare Diseases Center, West China Hospital, Sichuan University, Chengdu, Sichuan 610041, China.