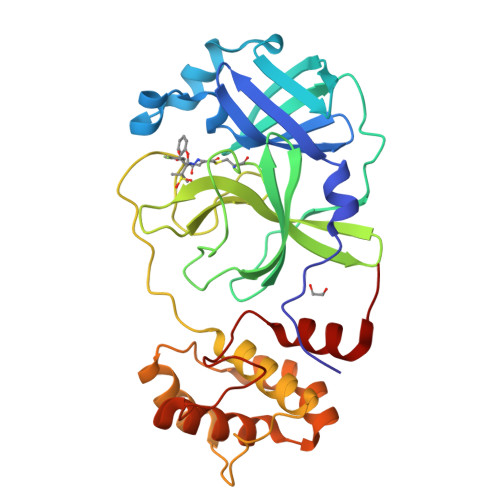
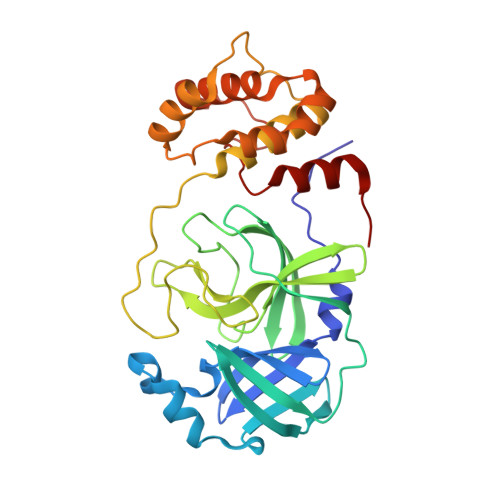
Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine.
Malla, T.R., Brewitz, L., Muntean, D.G., Aslam, H., Owen, C.D., Salah, E., Tumber, A., Lukacik, P., Strain-Damerell, C., Mikolajek, H., Walsh, M.A., Schofield, C.J.(2022) J Med Chem 65: 7682-7696
- PubMed: 35549342
- DOI: https://doi.org/10.1021/acs.jmedchem.1c02214
- Primary Citation of Related Structures:
7Z59 - PubMed Abstract:
The SARS-CoV-2 main protease (M pro ) is a medicinal chemistry target for COVID-19 treatment. Given the clinical efficacy of β-lactams as inhibitors of bacterial nucleophilic enzymes, they are of interest as inhibitors of viral nucleophilic serine and cysteine proteases. We describe the synthesis of penicillin derivatives which are potent M pro inhibitors and investigate their mechanism of inhibition using mass spectrometric and crystallographic analyses. The results suggest that β-lactams have considerable potential as M pro inhibitors via a mechanism involving reaction with the nucleophilic cysteine to form a stable acyl-enzyme complex as shown by crystallographic analysis. The results highlight the potential for inhibition of viral proteases employing nucleophilic catalysis by β-lactams and related acylating agents.
Organizational Affiliation:
Chemistry Research Laboratory, Department of Chemistry and the Ineos Oxford Institute for Antimicrobial Research, University of Oxford, 12 Mansfield Road, OX1 3TA Oxford, United Kingdom.