Synthesis of a Thiazole Library via an Iridium-Catalyzed Sulfur Ylide Insertion Reaction.
Hassell-Hart, S., Speranzini, E., Srikwanjai, S., Hossack, E., Roe, S.M., Fearon, D., Akinbosede, D., Hare, S., Spencer, J.(2022) Org Lett 24: 7924-7927
- PubMed: 36265082
- DOI: https://doi.org/10.1021/acs.orglett.2c02996
- Primary Citation of Related Structures:
8AXP - PubMed Abstract:
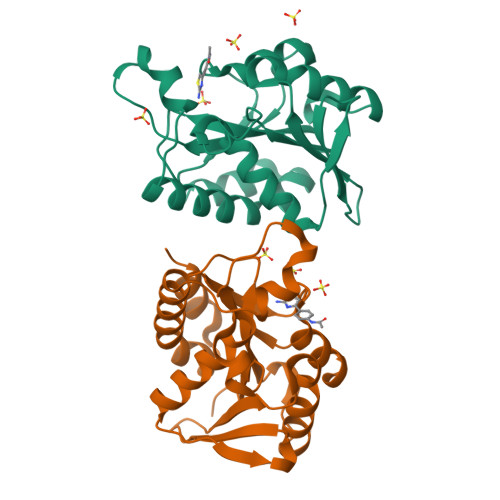
A library of thiazoles and selenothiazoles were synthesized via Ir-catalyzed ylide insertion chemistry. This process is a functional group, particularly heterocycle-substituent tolerant. This was applied to the synthesis of fanetizole, an anti-inflammatory drug, and a thiazole-containing drug fragment that binds to the peptidyl-tRNA hydrolase (Pth) in Neisseria gonorrheae bacteria.
Organizational Affiliation:
Department of Chemistry, School of Life Sciences, University of Sussex, Brighton BN1 9QJ, U.K.