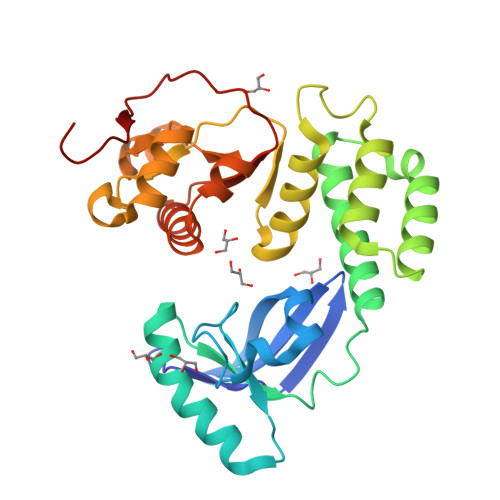
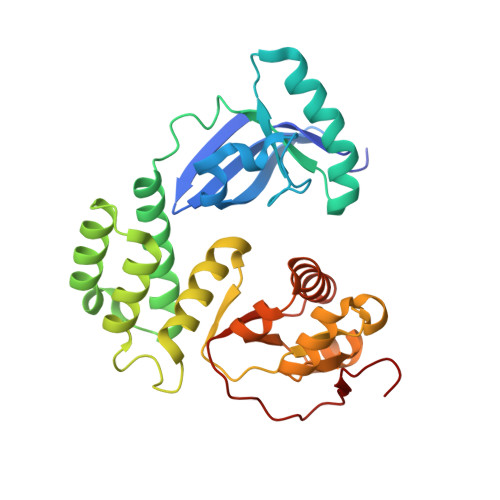
Crystal structure of the CoV-Y domain of SARS-CoV-2 nonstructural protein 3.
Li, Y., Pustovalova, Y., Shi, W., Gorbatyuk, O., Sreeramulu, S., Schwalbe, H., Hoch, J.C., Hao, B.(2023) Sci Rep 13: 2890-2890
- PubMed: 36801935
- DOI: https://doi.org/10.1038/s41598-023-30045-9
- Primary Citation of Related Structures:
8F2E - PubMed Abstract:
Replication of the coronavirus genome starts with the formation of viral RNA-containing double-membrane vesicles (DMV) following viral entry into the host cell. The multi-domain nonstructural protein 3 (nsp3) is the largest protein encoded by the known coronavirus genome and serves as a central component of the viral replication and transcription machinery. Previous studies demonstrated that the highly-conserved C-terminal region of nsp3 is essential for subcellular membrane rearrangement, yet the underlying mechanisms remain elusive. Here we report the crystal structure of the CoV-Y domain, the most C-terminal domain of the SARS-CoV-2 nsp3, at 2.4 Å-resolution. CoV-Y adopts a previously uncharacterized V-shaped fold featuring three distinct subdomains. Sequence alignment and structure prediction suggest that this fold is likely shared by the CoV-Y domains from closely related nsp3 homologs. NMR-based fragment screening combined with molecular docking identifies surface cavities in CoV-Y for interaction with potential ligands and other nsps. These studies provide the first structural view on a complete nsp3 CoV-Y domain, and the molecular framework for understanding the architecture, assembly and function of the nsp3 C-terminal domains in coronavirus replication. Our work illuminates nsp3 as a potential target for therapeutic interventions to aid in the on-going battle against the COVID-19 pandemic and diseases caused by other coronaviruses.
Organizational Affiliation:
Department of Molecular Biology and Biophysics, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, CT, 06030-3305, USA.