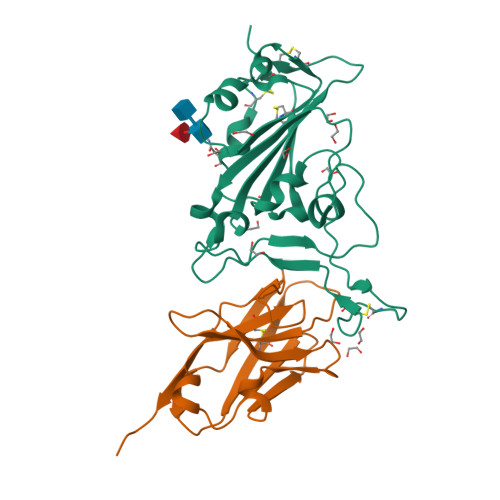
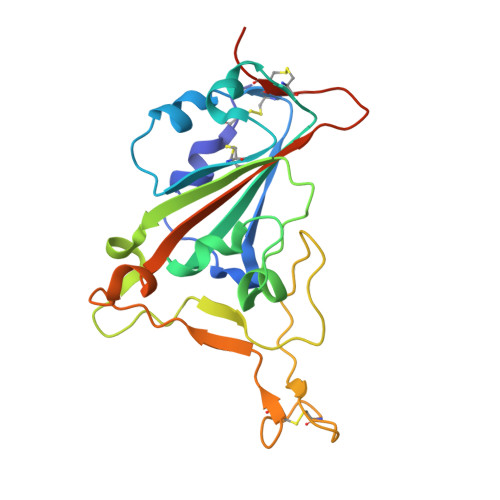
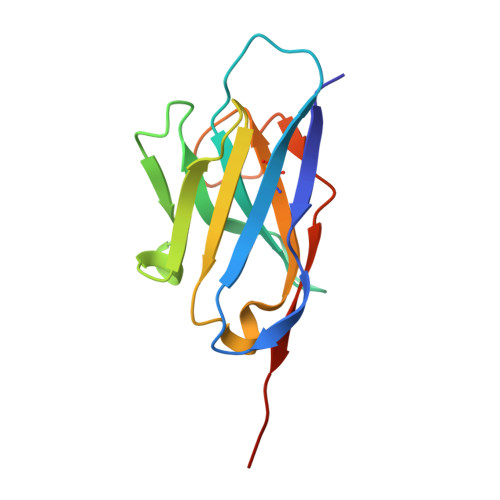
Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
Yamaguchi, K., Anzai, I., Maeda, R., Moriguchi, M., Watanabe, T., Imura, A., Takaori-Kondo, A., Inoue, T.(2023) J Biochem 173: 115-127
- PubMed: 36413757
- DOI: https://doi.org/10.1093/jb/mvac096
- Primary Citation of Related Structures:
8GZ5, 8GZ6 - PubMed Abstract:
The continuous emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants associated with the adaptive evolution of the virus is prolonging the global coronavirus disease 2019 (COVID-19) pandemic. The modification of neutralizing antibodies based on structural information is expected to be a useful approach to rapidly combat emerging variants. A dimerized variable domain of heavy chain of heavy chain antibody (VHH) P17 that has highly potent neutralizing activity against SARS-CoV-2 has been reported but the mode of interaction with the epitope remains unclear. Here, we report the X-ray crystal structure of the complex of monomerized P17 bound to the SARS-CoV-2 receptor binding domain (RBD) and investigated the binding activity of P17 toward various variants of concern (VOCs) using kinetics measurements. The structure revealed details of the binding interface and showed that P17 had an appropriate linker length to have an avidity effect and recognize a wide range of RBD orientations. Furthermore, we identified mutations in known VOCs that decrease the binding affinity of P17 and proposed methods for the acquisition of affinity toward the Omicron RBD because Omicron is currently the most predominant VOC. This study provides information for the rational design of effective VHHs for emerging VOCs.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, Osaka University, 1-6 Yamada-oka, Suita, Osaka 565-0871, Japan.