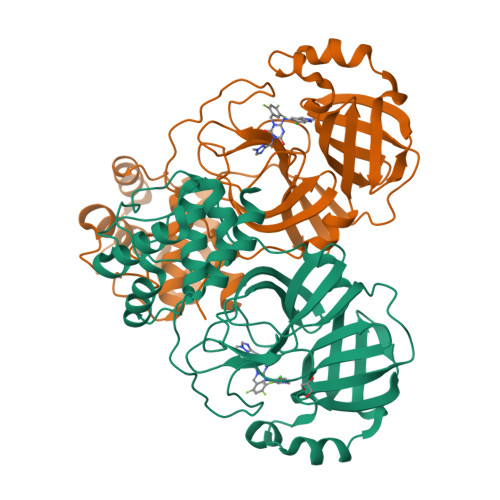
Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Yang, Y., Cao, L., Yan, M., Zhou, J., Yang, S., Xu, T., Huang, S., Li, K., Zhou, Q., Li, G., Zhu, Y., Cong, F., Zhang, H., Guo, D., Li, Y., Zhang, X.(2023) Antiviral Res 213: 105586-105586
- PubMed: 36997073
- DOI: https://doi.org/10.1016/j.antiviral.2023.105586
- Primary Citation of Related Structures:
8HEF - PubMed Abstract:
S-217622 (Ensitrelvir) is a reversible severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) 3-chymotrypsin-like protease (3CL pro ) inhibitor which obtained emergency regulatory approval in Japan for the treatment of SARS-CoV-2 infection on Nov 22, 2022. Herein, analogs of S-271622 with deuterium-for-hydrogen replacement were synthesized for comparison of the antiviral activities and pharmacokinetic (PK) profiles. Compared to the parent compound, C11-d2-S-217622 compound YY-278 retained in vitro activity against 3CL pro and SARS-CoV-2. X-ray crystal structural studies showed similar interactions of SARS-CoV-2 3CL pro with YY-278 and S-271622. The PK profiling revealed the relatively favorable bioavailability and plasma exposure of YY-278. In addition, YY-278, as well as S-217622, displayed broadly anti-coronaviral activities against 6 other coronaviruses that infect humans and animals. These results laid the foundation for further research on the therapeutic potential of YY-278 against COVID-19 and other coronaviral diseases.
Organizational Affiliation:
Department of Chemistry, College of Science, Academy for Advanced Interdisciplinary Studies, and Medi-X Pingshan, Southern University of Science and Technology, Shenzhen, Guangdong, 518000, China.