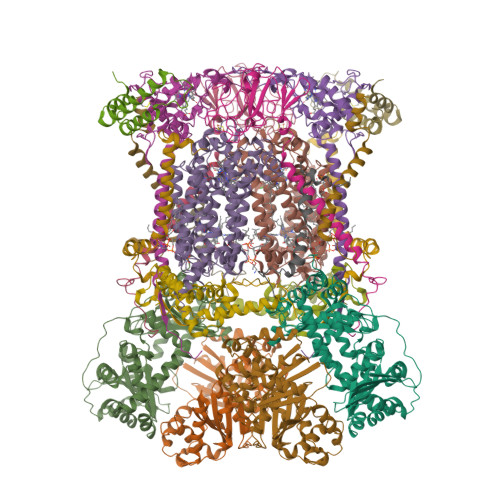
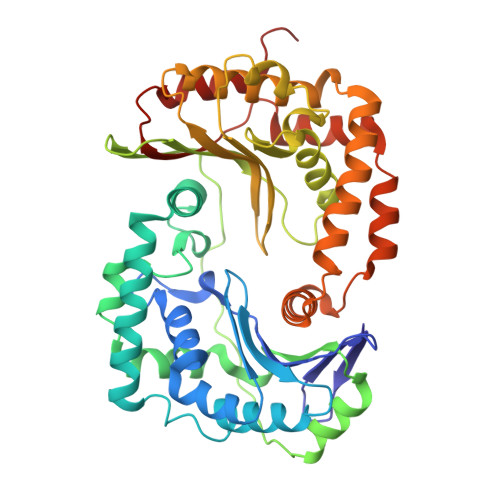
Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
Wang, Y.X., Ye, Y., Li, Z.W., Cui, G.R., Shi, X.X., Dong, Y., Jiang, J.J., Sun, J.Y., Guan, Z.W., Zhang, N., Wu, Q.Y., Wang, F., Zhu, X.L., Yang, G.F.(2024) J Am Chem Soc 146: 33903-33913
- PubMed: 39601138
- DOI: https://doi.org/10.1021/jacs.4c12595
- Primary Citation of Related Structures:
8YHQ, 8YIN, 8ZMT, 8ZOS, 8ZOW, 8ZP0 - PubMed Abstract:
Cytochrome bc 1 (complex III) represents a significant target for the discovery of both drugs and fungicides. Metyltetraprole (MET) is commonly classified as a quinone site inhibitor (Q o I) that combats the G143A mutated isolate, which confers high resistance to strobilurin fungicides such as pyraclostrobin (PYR). The binding mode and antiresistance mechanism of MET remain unclear. Here, we determined the high-resolution structures of inhibitor-bound S. cerevisiae complex III (MET, 2.52 Å; PYR, 2.42 Å) and inhibitor-bound porcine complex III (MET, 2.53 Å; PYR, 2,37 Å) by cryo-electron microscopy. The distinct binding modes of MET and PYR were observed for the first time. Notably, the MET exhibited different binding modes in the two species. In S. cerevisiae , the binding site of MET was the same as PYR, serving as a Pm -type inhibitor of the Q o site. However, in porcine, MET acted as a dual-target inhibitor of both Q o and Q i . Based on the structural insights, a novel inhibitor (YF23694) was discovered and demonstrated excellent fungicidal activity against downy mildew and powdery mildew fungi. This work provides a new starting point for the design of the next generation of inhibitors to overcome the resistance.
Organizational Affiliation:
State Key Laboratory of Green Pesticide, International Joint Research Center for Intelligent Biosensor Technology and Health, Central China Normal University, Wuhan 430079, PR China.