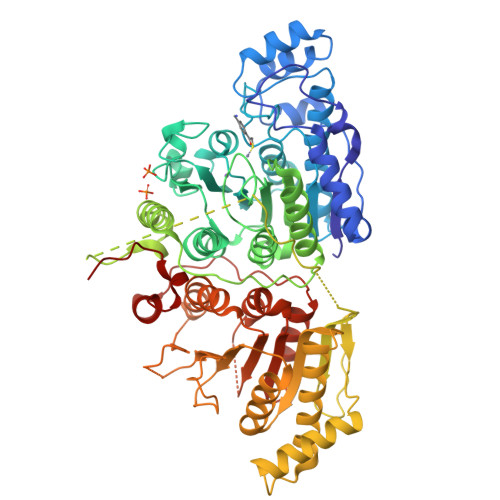
Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10.
Goulart Stollmaier, J., Watson, P.R., Christianson, D.W.(2024) ACS Med Chem Lett 15: 1715-1723
- PubMed: 39411528
- DOI: https://doi.org/10.1021/acsmedchemlett.4c00293
- Primary Citation of Related Structures:
9CBF, 9CBG, 9CBH, 9CBI, 9CBJ, 9CBK - PubMed Abstract:
Histone deacetylase 10 (HDAC10) is unique among the greater HDAC family due to its unusually narrow substrate specificity as a polyamine deacetylase, specifically as an N 8 -acetylspermidine hydrolase. Polyamines are essential for cell growth and proliferation; consequently, inhibition of polyamine deacetylation represents a possible strategy for cancer chemotherapy. In this work, we have designed six acetylated phenylthioketone inhibitors of HDAC10 containing positively charged para - and meta -substituted amino groups designed to target interactions with E274, the gatekeeper that recognizes the positively charged ammonium group of the substrate N 8 -acetylspermidine. We prepared each of these inhibitors through a short synthetic route of six steps. By adapting a low-cost colorimetric activity assay, we measured low-micromolar IC 50 values for these compounds against a humanized construct of zebrafish HDAC10 (A24E-D94A HDAC10). Selected inhibitors were cocrystallized with A24E-D94A zebrafish HDAC10 and zebrafish HDAC6 to provide insight into class IIb isozyme affinity and selectivity.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104-6323, United States.