Full Text |
QUERY: Citation Author = "De Carlo, S." | MyPDB Login | Search API |
| Search Summary | This query matches 7 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 7 of 7 Structures Page 1 of 1 Sort by
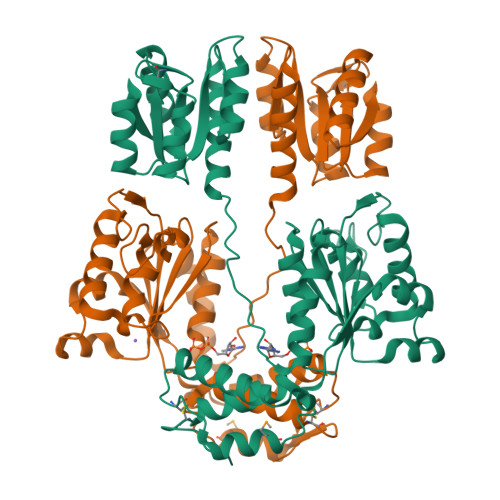
Crystal structure of sigma54 activator NTRC4 in the inactive stateBatchelor, J.D., Doucleff, M., Lee, C.-J., Matsubara, K., De Carlo, S., Heideker, J., Lamers, M.M., Pelton, J.G., Wemmer, D.E. (2008) J Mol Biology 384: 1058-1075
Crystal structure of sigma54 activator NtrC4's DNA binding domainBatchelor, J.D., Doucleff, M., Lee, C.-J., Matsubara, K., De Carlo, S., Heideker, J., Lamers, M.M., Pelton, J.G., Wemmer, D.E. (2008) J Mol Biology 384: 1058-1075
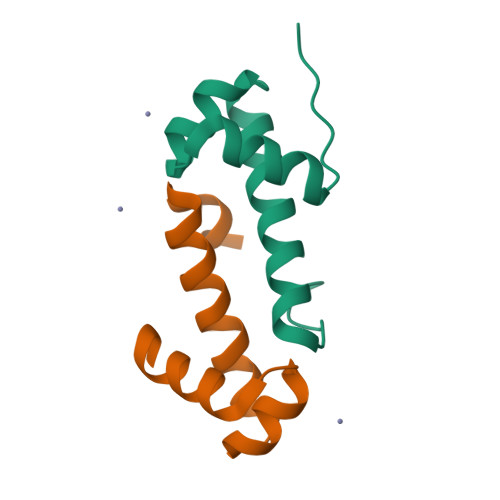
Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domainChen, B., Sysoeva, T.A., Chowdhury, S., Rusu, M., Birmanns, S., Guo, L., Hanson, J., Yang, H., Nixon, B.T. (2010) Structure 18: 1420-1430
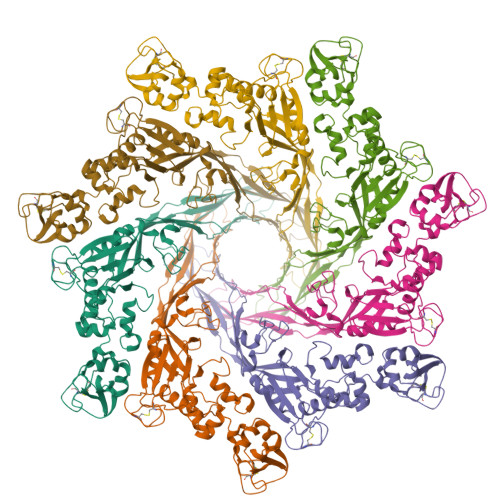
Cryo-EM structure of aerolysin prepore(2016) Nat Commun 7: 12062-12062
Cryo-EM structure of aerolysin pore in LMNG micelle(2016) Nat Commun 7: 12062-12062
Cryo-EM structures of aerolysin post-prepore and quasipore(2016) Nat Commun 7: 12062-12062
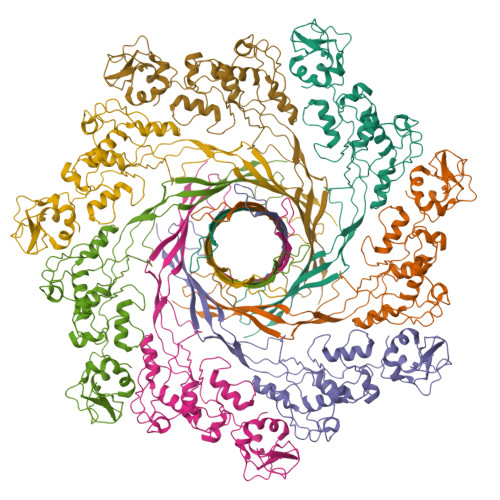
Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris hemoglobinAfanasyev, P., Linnemayr-Seer, C., Ravelli, R.B.G., Matadeen, R., De Carlo, S., Alewijnse, B., Portugal, R.V., Pannu, N.S., Schatz, M., van Heel, M. (2017) IUCrJ 4: 678-694
1 to 7 of 7 Structures Page 1 of 1 Sort by |