Full Text |
QUERY: Citation Author = "Roche, J." | MyPDB Login | Search API |
| Search Summary | This query matches 37 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismMore... TaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 25 of 37 Structures Page 1 of 2 Sort by
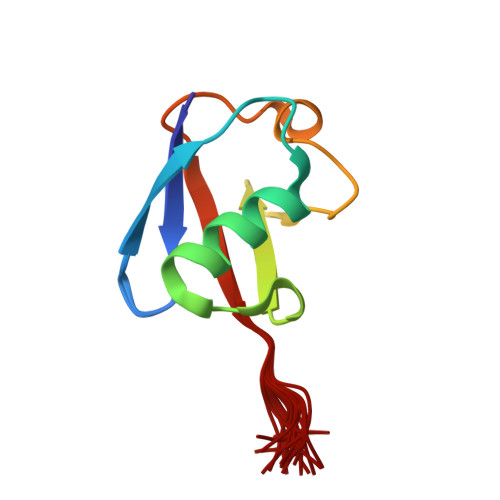
Solution nmr structure of ubiquitin refined against dipolar couplings in 4 mediaMaltsev, A., Grishaev, A., Roche, J., Zasloff, M., Bax, A. (2014) J Am Chem Soc 136: 3752-3755
Solution NMR structure of gp41 ectodomain monomer on a DPC micelleRoche, J., Louis, J.M., Grishaev, A., Ying, J., Bax, A. (2014) Proc Natl Acad Sci U S A 111: 3425-3430
Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS L38A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS F34A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS L103A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS V74A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno E., B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS L36A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS L125A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A at cryogenic temperatureCaro, J.A., Schlessman, J.L., Garcia-Moreno, E.B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS V23A at cryogenic temperatureCaro, J.A., Clark, I., Schlessman, J.L., Garcia-Moreno E., B., Heroux, A. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperature and with high redundancyCaro, J.A., Schlessman, J.L., Garcia-Moreno E., B. (2012) Proc Natl Acad Sci U S A 109: 6945-6950
Llama nanobody PorM_130(2017) Acta Crystallogr F Struct Biol Commun 73: 286-293
Llama nanobody PorM_19Leone, P., Cambillau, C., Roussel, A. (2017) Acta Crystallogr F Struct Biol Commun 73: 286-293
Llama nanobody PorM_02Roche, J., Gaubert, A., Leone, P., Roussel, A. (2017) Acta Crystallogr F Struct Biol Commun 73: 286-293
Llama nanobody PorM_01Duhoo, Y., Leone, P., Roussel, A. (2017) Acta Crystallogr F Struct Biol Commun 73: 286-293
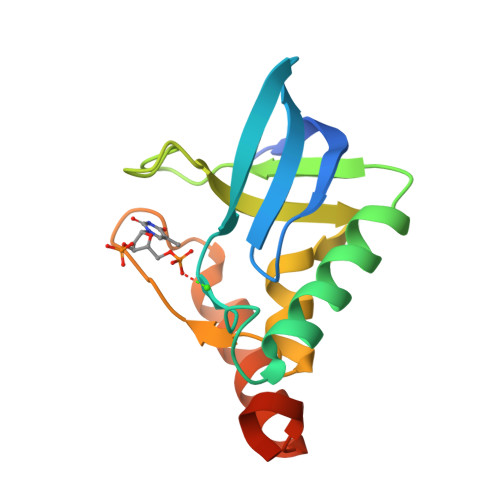
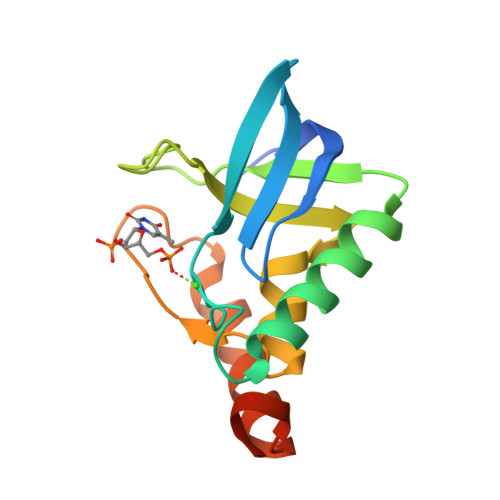
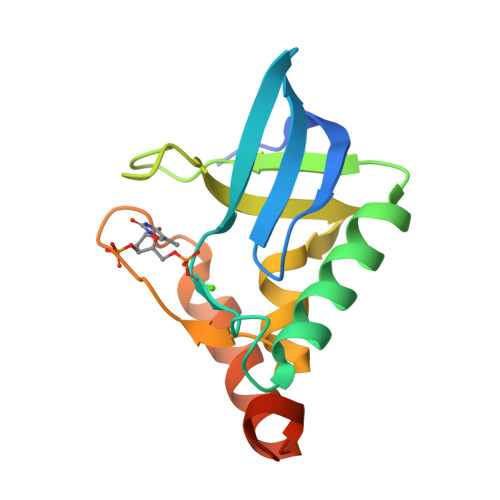
Crystal Structure of multi-drug resistant HIV-1 protease PR-S17(2016) PLoS One 11: e0168616-e0168616
Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir(2016) PLoS One 11: e0168616-e0168616
N-terminal part (residues 30-212) of PorM with the llama nanobody nb01Leone, P., Roche, J., Cambillau, C., Roussel, A. (2018) Nat Commun 9: 429-429
Periplasmic domain (residues 36-513) of GldMLeone, P., Roche, J., Cambillau, C., Roussel, A. (2018) Nat Commun 9: 429-429
C-terminal part (residues 224-515) of PorMLeone, P., Cambillau, C., Roussel, A. (2018) Nat Commun 9: 429-429
C-terminal part (residues 315-516) of PorM with the llama nanobody nb130Leone, P., Cambillau, C., Roussel, A. (2018) Nat Commun 9: 429-429
Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic deviceRoche, J., Gaubert, A., Desmyter, A., De Wijn, R., Sauter, C., Roussel, A. (2019) IUCrJ 6: 454-464
1 to 25 of 37 Structures Page 1 of 2 Sort by |