Full Text |
QUERY: Citation Author = "Trautwein-Schult, A." | MyPDB Login | Search API |
| Search Summary | This query matches 5 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 5 of 5 Structures Page 1 of 1 Sort by
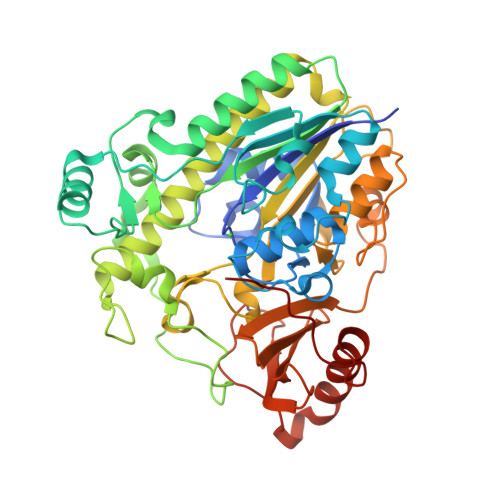
Crystal structure of the family S1_7 ulvan-specific sulfatase FA22070 from Formosa agariphilaRoret, T., Prechoux, A., Michel, G., Czjzek, M. (2019) Nat Chem Biol 15: 803-812
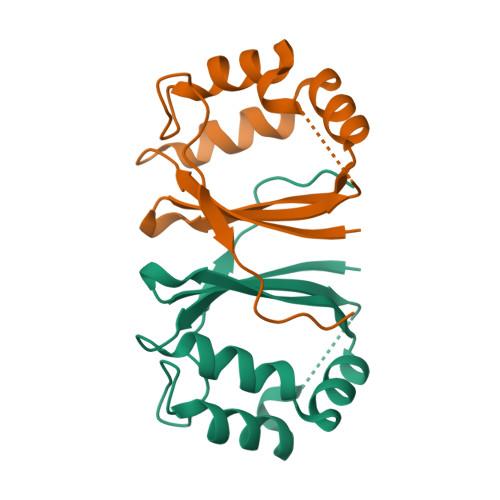
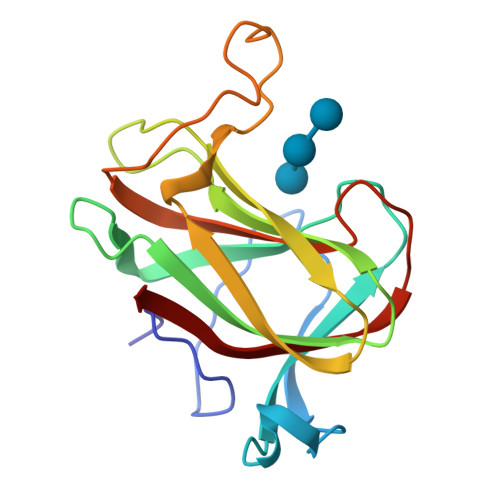
Crystal structure of L-rhamnose mutarotase FA22100 from Formosa agariphilaRoret, T., Prechoux, A., Michel, G., Czjzek, M. (2019) Nat Chem Biol 15: 803-812
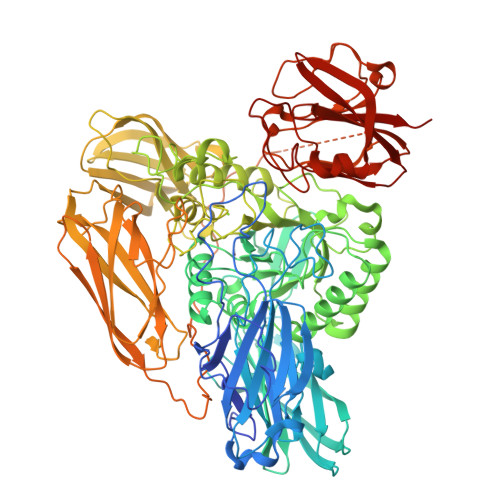
The structure of a beta-glucuronidase from glycoside hydrolase family 2Robb, C.S., Gerlach, N., Reisky, L., Bornshoeru, U., Hehemann, J.H. (2019) Nat Chem Biol 15: 803-812
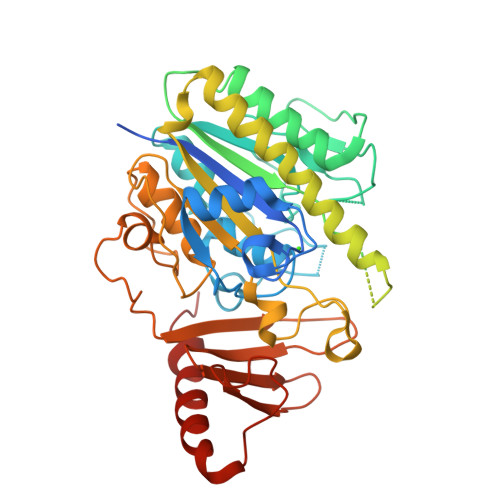
Structure of the S1_25 family sulfatase module of the rhamnosidase FA22250 from Formosa agariphilaRoret, T., Prechoux, A., Czjzek, M., Michel, G. (2019) Nat Chem Biol 15: 803-812
Novel laminarin-binding CBM X584Zuehlke, M.K., Jeudy, A., Czjzek, M. (2024) Environ Microbiol 26: e16624-e16624
1 to 5 of 5 Structures Page 1 of 1 Sort by |