Full Text |
QUERY: Gene Name = "SIR3" | MyPDB Login | Search API |
| Search Summary | This query matches 10 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateSymmetry TypeSCOP Classification | 1 to 10 of 10 Structures Page 1 of 1 Sort by
S. cerevisiae Sir3 BAH domainKeck, J.L., Hou, Z., Daner, J.R., Fox, C.A. (2006) Protein Sci 15: 1182-1186
Structure of the yeast Sir3 BAH domain(2006) Mol Cell Biol 26: 3256-3265
Crystal structure of S. cerevisiae RAP1-Sir3 complex(2011) Nat Struct Mol Biol 18: 213-221
Crystal Structure of the S. cerevisiae Sir3 AAA+ domain(2011) Genes Dev 25: 1835-1846
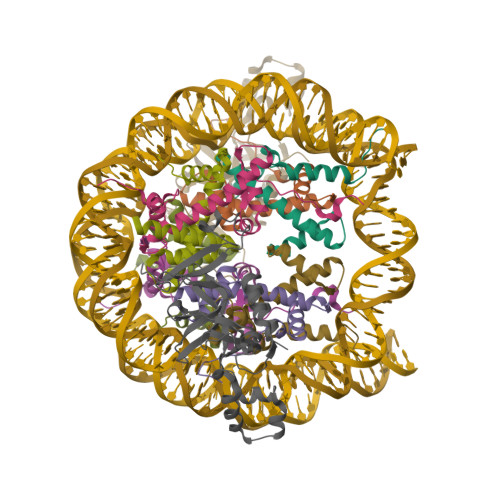
Crystal structure of the Sir3 BAH domain in complex with a nucleosome core particle.Armache, K.-J., Garlick, J.D., Canzio, D., Narlikar, G.J., Kingston, R.E. (2011) Science 334: 977-982
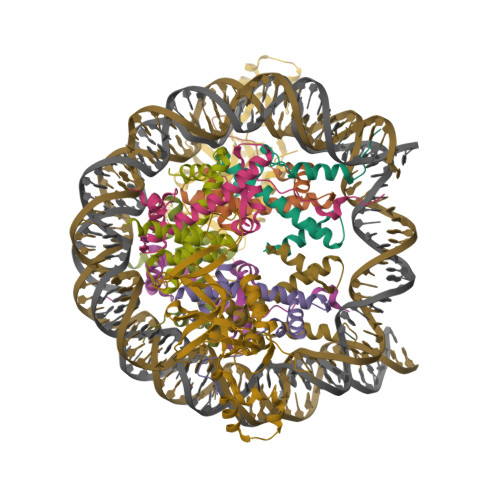
Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosomeWang, F., Li, G., Mohammed, A., Lu, C., Currie, M., Johnson, A., Moazed, D. (2013) Proc Natl Acad Sci U S A 110: 8495-8500
Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particleYang, D., Fang, Q., Wang, M., Ren, R., Wang, H., He, M., Sun, Y., Yang, N., Xu, R.M. (2013) Nat Struct Mol Biol 20: 1116-1118
Crystal structure of N-terminal acetylated yeast Sir3 BAH domainYang, D., Fang, Q., Wang, M., Ren, R., Wang, H., He, M., Sun, Y., Yang, N., Xu, R.M. (2013) Nat Struct Mol Biol 20: 1116-1118
Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutantYang, D., Fang, Q., Wang, M., Ren, R., Wang, H., He, M., Sun, Y., Yang, N., Xu, R.M. (2013) Nat Struct Mol Biol 20: 1116-1118
Crystal structure of the N-terminally acetylated BAH domain of Sir3 bound to the nucleosome core particleArnaudo, N., Fernandez, I.S., McLaughlin, S.H., Peak-Chew, S.Y., Rhodes, D., Martino, F. (2013) Nat Struct Mol Biol 20: 1119-1121
1 to 10 of 10 Structures Page 1 of 1 Sort by |