Full Text |
QUERY: Gene Name = "mecB" | MyPDB Login | Search API |
| Search Summary | This query matches 8 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateSymmetry Type | 1 to 8 of 8 Structures Page 1 of 1 Sort by
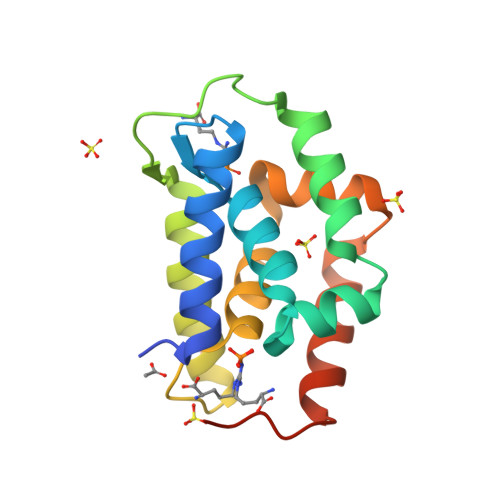
NMR solution structure of the Bacillus subtilis ClpC N-domainKojetin, D.J., McLaughlin, P.D., Thompson, R.J., Rance, M., Cavanagh, J. (2009) J Mol Biology 387: 639-652
Crystal Structure of the ATP-dependent Clp Protease ClpC from Clostridium difficileKim, Y., Tesar, C., Li, H., Cobb, G., Joachimiak, A., Midwest Center for Structural Genomics (MCSG) To be published
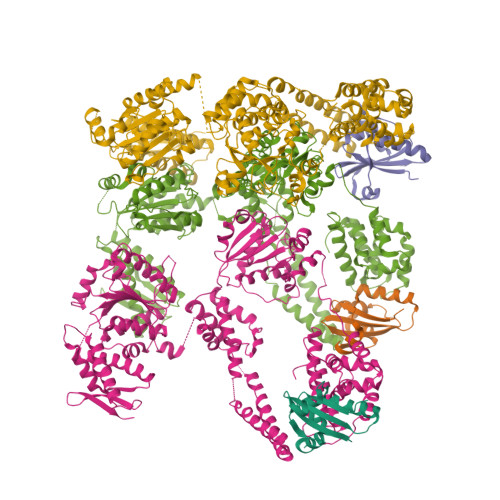
Structural dynamics of the MecA-ClpC complex revealed by cryo-EMLiu, J., Mei, Z., Li, N., Qi, Y., Xu, Y., Shi, Y., Wang, F., Lei, J., Gao, N. (2013) J Biological Chem 288: 17597-17608
Structural dynamics of the MecA-ClpC complex revealed by cryo-EMLiu, J., Mei, Z., Li, N., Qi, Y., Xu, Y., Shi, Y., Wang, F., Lei, J., Gao, N. (2013) J Biological Chem 288: 17597-17608
Structural dynamics of the MecA-ClpC complex revealed by cryo-EMLiu, J., Mei, Z., Li, N., Qi, Y., Xu, Y., Shi, Y., Wang, F., Lei, J., Gao, N. (2013) J Biological Chem 288: 17597-17608
Structure of MecA108:ClpCWang, F., Mei, Z.Q., Wang, J.W., Shi, Y.G. (2011) Nature 471: 331-335
ClpC N-terminal domain with bound phospho-arginine(2016) Nature 539: 48-53
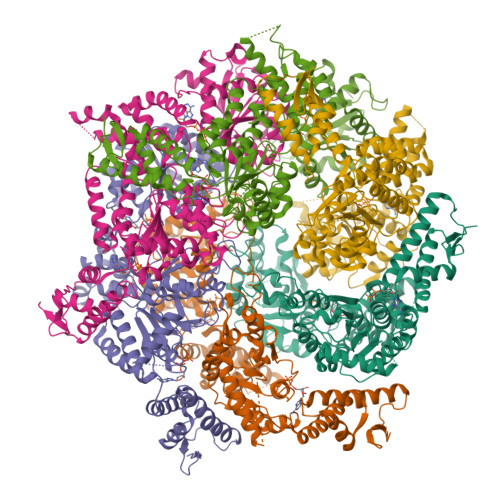
Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptideMorreale, F.E., Meinhart, A., Haselbach, D., Clausen, T. (2022) Cell 185: 2338
1 to 8 of 8 Structures Page 1 of 1 Sort by |