Full Text |
QUERY: Structure Keywords HAS EXACT PHRASE "TRANSCRIPTION/HYDROLASE" | MyPDB Login | Search API |
| Search Summary | This query matches 9 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 9 of 9 Structures Page 1 of 1 Sort by
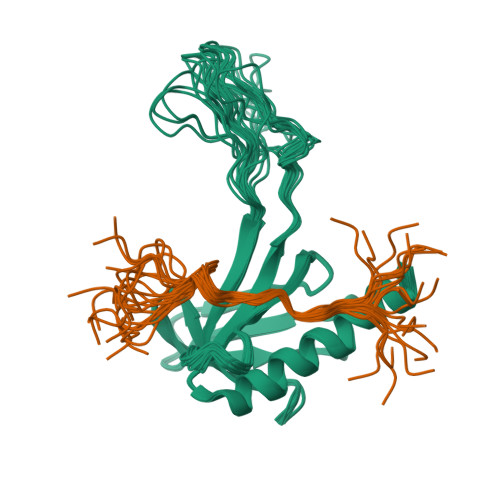
NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and Rad2Lafrance-Vanasse, J., Legault, P., Omichinski, J. (2012) Nucleic Acids Res 40: 5739-5750
Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4Hartmann, M.D., Djuranovic, S., Ursinus, A., Zeth, K., Lupas, A.N. (2009) Mol Cell 34: 580
Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A MutantHartmann, M.D., Djuranovic, S., Ursinus, A., Zeth, K., Lupas, A.N. (2009) Mol Cell 34: 580
Structure of yeast NOT1 MIF4G domain co-crystallized with CAF1(2012) Mol Cell 48: 207
Crystal Structure of MBP-fused Human Six1 Bound to Human Eya2 Eya Domain(2013) Nat Struct Mol Biol 20: 447-453
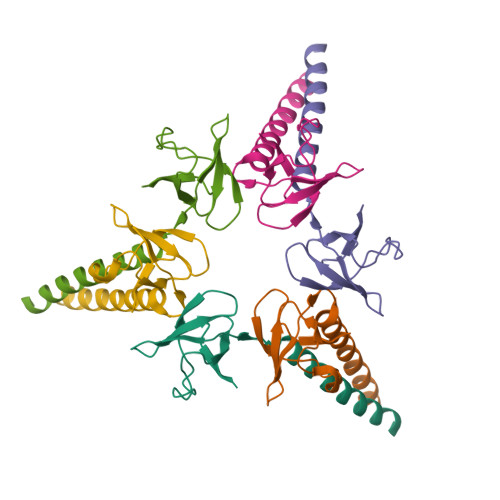
Structure of Arp7-Arp9-Snf2(HSA)-RTT102 subcomplex of SWI/SNF chromatin remodeler.Schubert, H.L., Cairns, B.R., Hill, C.P. (2013) Proc Natl Acad Sci U S A 110: 3345-3350
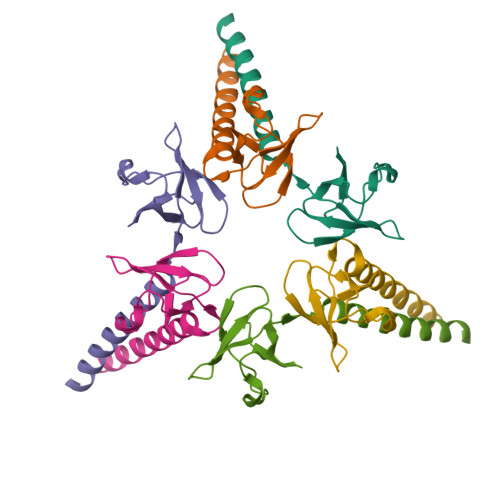
Structure of yeast SAGA DUBm with Sgf73 Y57A mutant at 2.8 angstroms resolution(2015) J Mol Biology 427: 1765-1778
Crystal structure of the E. coli transcription termination factor Rho(2020) Acta Crystallogr F Struct Biol Commun 76: 398-405
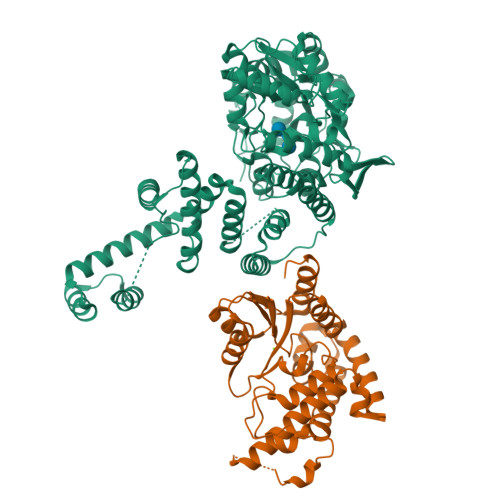
Crystal Structure Analysis of the Yeast Tyrosyl-DNA PhosphodiesteraseHe, X., Babaoglu, K., Price, A., Nitiss, K.C., Nitiss, J.L., White, S.W. (2007) J Mol Biology 372: 1070-1081
1 to 9 of 9 Structures Page 1 of 1 Sort by |