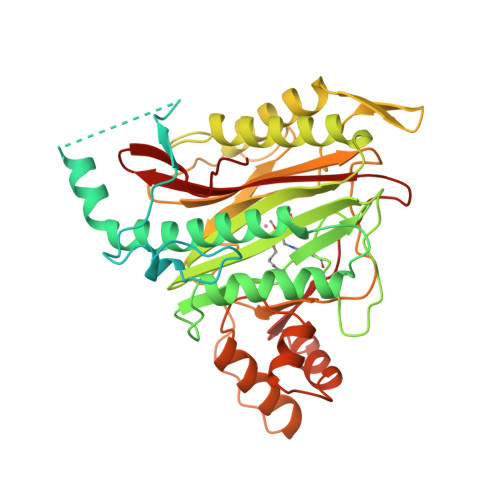
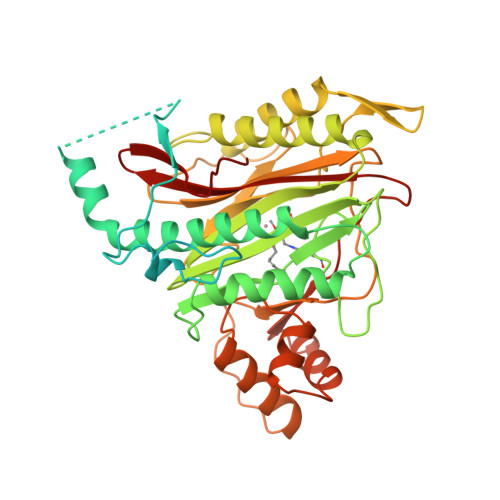
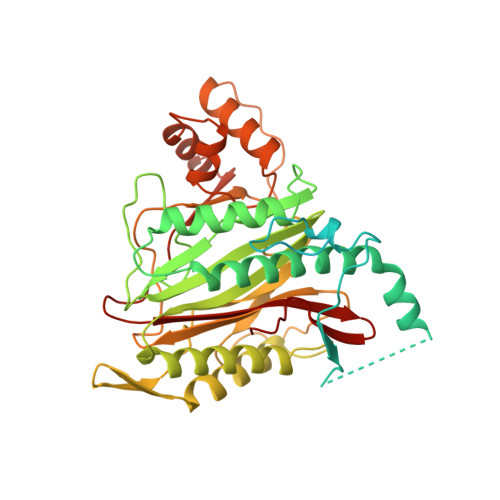
Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Liu, S., Widom, J., Kemp, C.W., Crews, C.M., Clardy, J.(1998) Science 282: 1324-1327
- PubMed: 9812898
- DOI: https://doi.org/10.1126/science.282.5392.1324
- Primary Citation of Related Structures:
1B59, 1B6A, 1BN5, 1BOA - PubMed Abstract:
The fungal metabolite fumagillin suppresses the formation of new blood vessels, and a fumagillin analog is currently in clinical trials as an anticancer agent. The molecular target of fumagillin is methionine aminopeptidase-2 (MetAP-2). A 1.8 A resolution crystal structure of free and inhibited human MetAP-2 shows a covalent bond formed between a reactive epoxide of fumagillin and histidine-231 in the active site of MetAP-2. Extensive hydrophobic and water-mediated polar interactions with other parts of fumagillin provide additional affinity. Fumagillin-based drugs inhibit MetAP-2 but not MetAP-1, and the three-dimensional structure also indicates the likely determinants of this specificity. The structural basis for fumagillin's potency and specificity forms the starting point for structure-based drug design.
Organizational Affiliation:
J. Clardy, Department of Chemistry and Chemical Biology, Baker Laboratory, Cornell University, Ithaca, NY 14853-1301, USA.