Structural interactions of fibroblast growth factor receptor with its ligands.
Stauber, D.J., DiGabriele, A.D., Hendrickson, W.A.(2000) Proc Natl Acad Sci U S A 97: 49-54
- PubMed: 10618369
- DOI: https://doi.org/10.1073/pnas.97.1.49
- Primary Citation of Related Structures:
1DJS - PubMed Abstract:
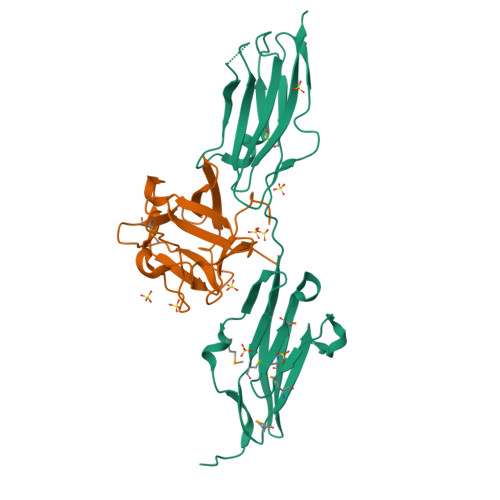
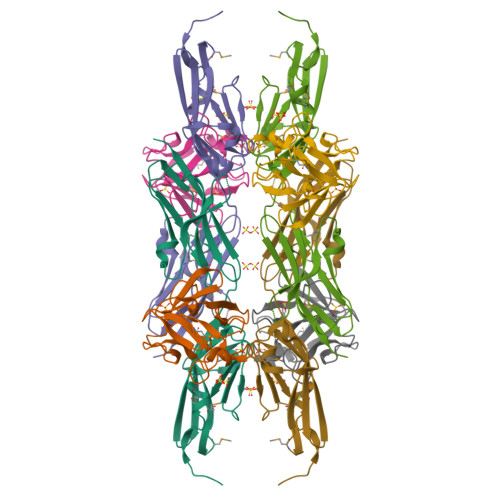
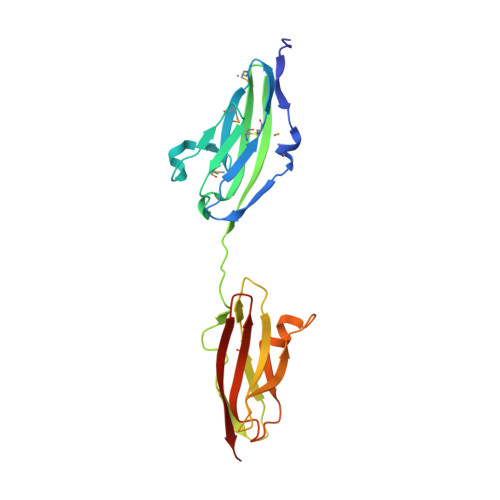
Fibroblast growth factors (FGFs) effect cellular responses by binding to FGF receptors (FGFRs). FGF bound to extracellular domains on the FGFR in the presence of heparin activates the cytoplasmic receptor tyrosine kinase through autophosphorylation. We have crystallized a complex between human FGF1 and a two-domain extracellular fragment of human FGFR2. The crystal structure, determined by multiwavelength anomalous diffraction analysis of the selenomethionyl protein, is a dimeric assemblage of 1:1 ligand:receptor complexes. FGF is bound at the junction between the two domains of one FGFR, and two such units are associated through receptor:receptor and secondary ligand:receptor interfaces. Sulfate ion positions appear to mark the course of heparin binding between FGF molecules through a basic region on receptor D2 domains. This dimeric assemblage provides a structural mechanism for FGF signal transduction.
Organizational Affiliation:
Department of Biochemistry, Columbia University, New York, NY 10032, USA.