Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
Airenne, T.T., Torkko, J.M., Van Der Plas, S., Sormunen, R.T., Kastaniotis, A.J., Wierenga, R.K., Hiltunen, J.K.(2003) J Mol Biology 327: 47
- PubMed: 12614607
- DOI: https://doi.org/10.1016/s0022-2836(03)00038-x
- Primary Citation of Related Structures:
1GU7, 1GUF, 1GYR - PubMed Abstract:
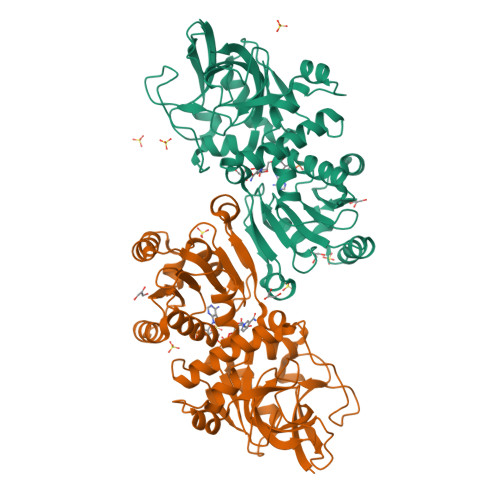
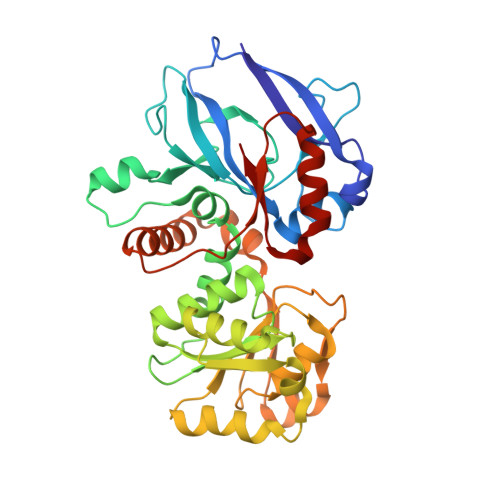
Candida tropicalis enoyl thioester reductase Etr1p and the Saccharomyces cerevisiae homologue Mrf1p catalyse the NADPH-dependent reduction of trans-2-enoyl thioesters in mitochondrial fatty acid synthesis (FAS). Unlike prokaryotic enoyl thioester reductases (ETRs), which belong to the short-chain dehydrogenases/reductases (SDR), Etr1p and Mrf1p represent structurally distinguishable ETRs that belong to the medium-chain dehydrogenases/reductases (MDR) superfamily, indicating independent origin of two separate classes of ETRs. The crystal structures of Etr1p, the Etr1p-NADPH complex and the Etr1Y79Np mutant were refined to 1.70A, 2.25A and 2.60A resolution, respectively. The native fold of Etr1p was maintained in Etr1Y79Np, but the mutant had only 0.1% of Etr1p catalytic activity remaining and failed to rescue the respiratory deficient phenotype of the mrf1Delta strain. Mutagenesis of Tyr73 in Mrf1p, corresponding to Tyr79 in Etr1p, produced similar results. Our data indicate that the mitochondrial reductase activity is indispensable for respiratory function in yeast, emphasizing the significance of Mrf1p (Etr1p) and mitochondrial FAS for the integrity of the respiratory competent organelle.
Organizational Affiliation:
Biocenter Oulu and Department of Biochemistry, P.O. Box 3000, FIN-90014 University of Oulu, Finland.