Axial ligand complexes of the Rhodnius nitrophorins: reduction potentials, binding constants, EPR spectra, and structures of the 4-iodopyrazole and imidazole complexes of NP4
Berry, R.E., Ding, X.D., Shokhireva, T.K., Weichsel, A., Montfort, W.R., Walker, F.A.(2004) J Biol Inorg Chem 9: 135-144
- PubMed: 14673714
- DOI: https://doi.org/10.1007/s00775-003-0505-0
- Primary Citation of Related Structures:
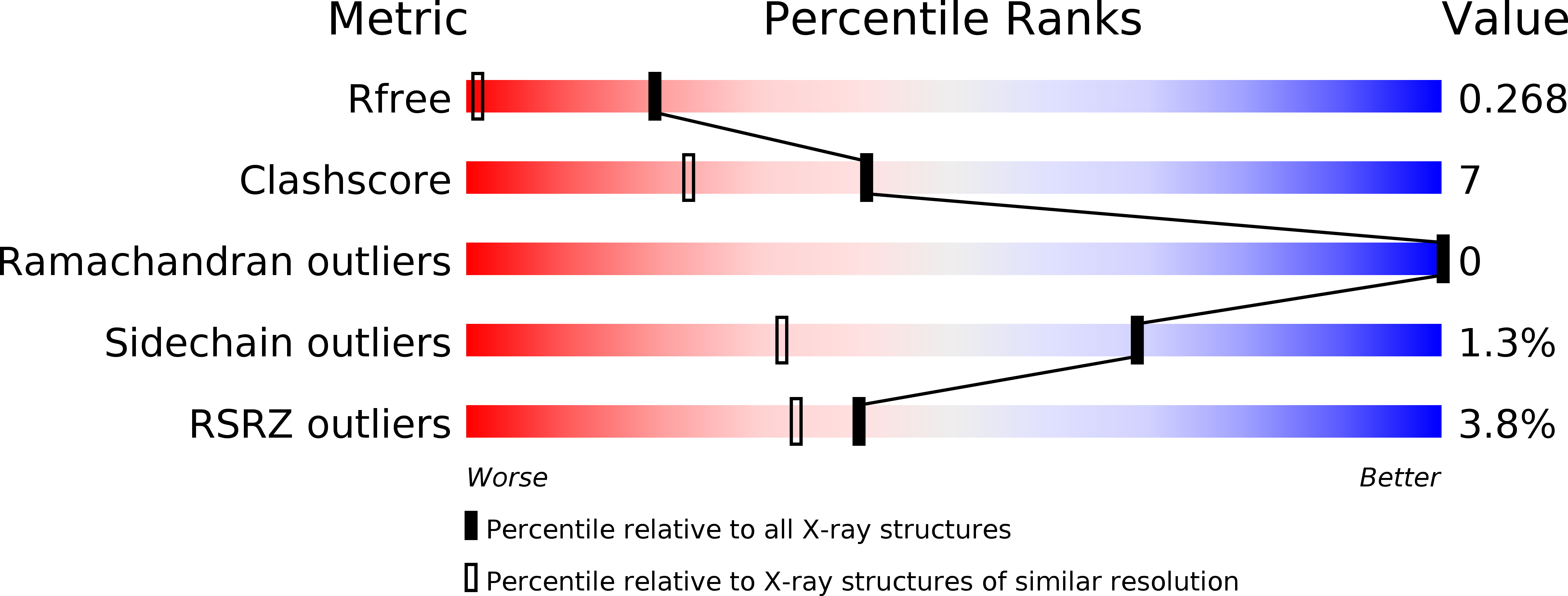
1ML7 - PubMed Abstract:
Previously, we utilized 4-iodopyrazole (4IPzH) as a heavy atom derivative for the initial solution of the crystal structure of the nitrophorin from Rhodnius prolixus, NP1, where it was found to bind to the heme with the iodo group disordered in two positions. We have now determined the structure of the 4IPzH complex of NP4 at pH 7.5 and find that the geometry and bond lengths at the iron center are extremely similar to those of the imidazole (ImH) complex of the same protein (structure determined at pH 5.6), except that the G-H loop is not in the closed conformation. 4IPzH binds to the heme of NP4 in an ordered manner, with the iodo substituent pointed toward the opening of the heme pocket, near the surface of the protein. In order to understand the solution chemistry in terms of the relative binding abilities of 4IPzH, ImH, and histamine (Hm, a physiological ligand for the nitrophorins), we have also investigated the equilibrium binding constants and reduction potentials of these three ligand complexes of the four Rhodnius nitrophorins as a function of pH. We have found that, unlike the other Lewis bases, 4IPzH forms less stable complexes with the Fe(III) than the Fe(II) oxidation states of NP1 and NP4, and similar stability for the two oxidation states of NP2 and NP3, suggesting that this ligand is a softer base than ImH or Hm, for both of which the Fe(III) complexes are more stable than those of Fe(II) for all four nitrophorins. Surprisingly, in spite of this and the much lower basicity of 4IPzH than imidazole and histamine, the EPR g-values of all three ligand complexes are very similar.
Organizational Affiliation:
Department of Chemistry, University of Arizona, Tucson, AZ 85721, USA.