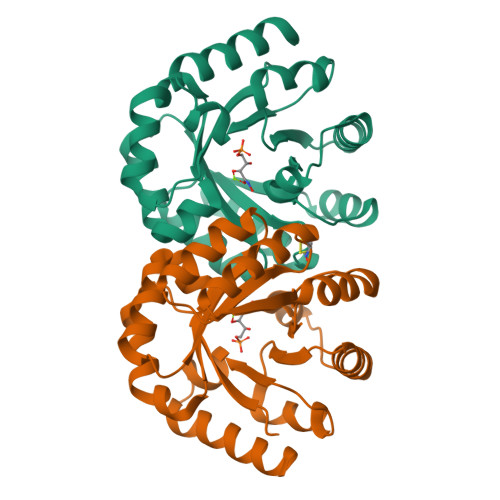
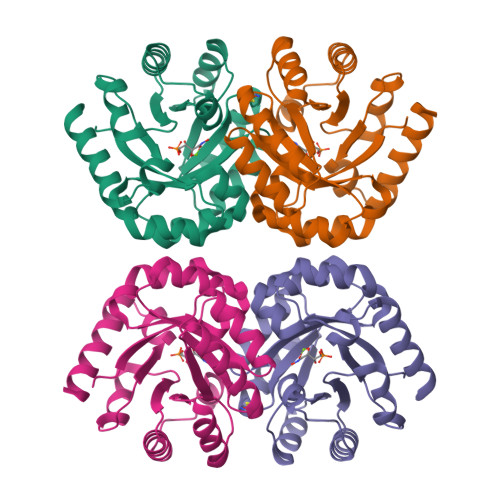
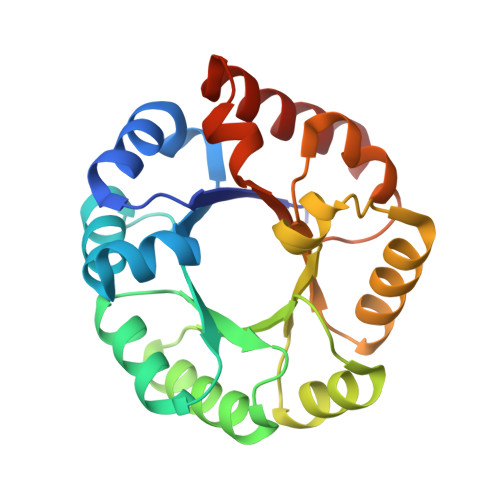
Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Wise, E.L., Yew, W.S., Gerlt, J.A., Rayment, I.(2003) Biochemistry 42: 12133-12142
- PubMed: 14567674
- DOI: https://doi.org/10.1021/bi0348819
- Primary Citation of Related Structures:
1Q6L, 1Q6O, 1Q6Q, 1Q6R - PubMed Abstract:
3-Keto-L-gulonate 6-phosphate decarboxylase (KGPDC) and orotidine 5'-phosphate decarboxylase (OMPDC) are members of an enzyme suprafamily, the OMPDC suprafamily, because they are homologous enzymes that catalyze mechanistically distinct reactions using different substrates. KGPDC catalyzes the Mg(2+) ion-dependent decarboxylation of 3-keto-L-gulonate 6-phosphate to yield L-xylulose 5-phosphate and CO(2); OMPDC catalyzes the metal ion-independent decarboxylation of OMP to UMP and CO(2). Structural studies have shown that KGPDC and OMPDC share several strictly conserved active site residues that are used differently by each enzyme to catalyze their mechanistically distinct reactions. Although the mechanism of the KGPDC-catalyzed reaction has yet to be elucidated, it is thought to proceed via a Mg(2+) ion-stabilized 1,2-enediolate intermediate. Here we report the crystal structures of KGPDC complexed with L-gulonate 6-phosphate, L-threonohydroxamate 4-phosphate, and L-xylitol 5-phosphate, analogues of the substrate, enediolate intermediate, and product, as well as with the product, L-xylulose 5-phosphate, at 1.2, 1.8, 1.7, and 1.8 A resolution, respectively. These structures support a mechanism that involves the formation of a cis-1,2-enediolate intermediate. Contrary to expectations, the geometry of the intermediate does not involve bidentate coordination of both enediolate oxygen atoms to the Mg(2+) ion but rather involves only the coordination of the oxygen on C2 to the Mg(2+) ion. The oxygen atom on C1 instead forms hydrogen bonds to both Lys64 and Asp67, two strictly conserved active site residues. Lys64 also interacts with the oxygen on C2 and may serve to stabilize a cis conformation of the 1,2-enediolate. These structures also implicate His136 to be the general acid that protonates the 1,2-enediolate intermediate. This study further demonstrates that multiple unrelated enzyme functions can evolve from a single active site architecture without regard for substrate binding affinity or mechanism.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706, USA.