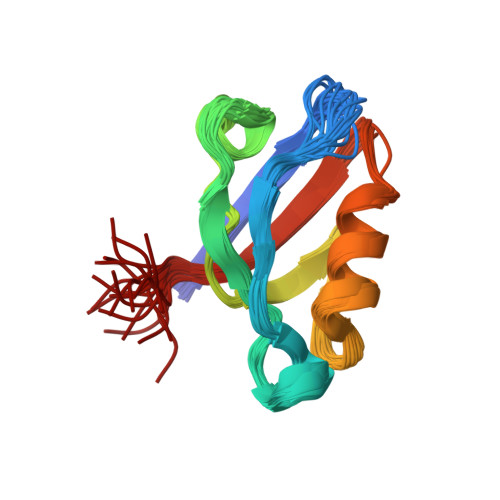
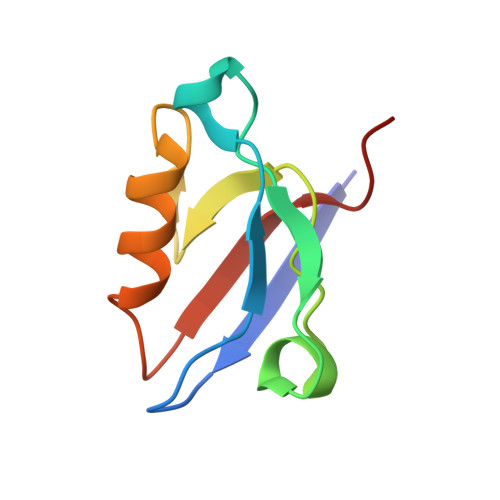
Solution Structure of ZASP PDZ Domain; Implications for Sarcomere Ultrastructure and Enigma Family Redundancy.
Au, Y., Atkinson, R.A., Guerrini, R., Kelly, G., Joseph, C., Martin, S.R., Muskett, F.W., Pallavicini, A., Faulkner, G., Pastore, A.(2004) Structure 12: 611-622
- PubMed: 15062084
- DOI: https://doi.org/10.1016/j.str.2004.02.019
- Primary Citation of Related Structures:
1RGW - PubMed Abstract:
Z band alternately spliced PDZ-containing protein (ZASP) is a sarcomere Z disk protein expressed in human cardiac and skeletal muscle that is thought to be involved in a dominant familial dilated cardiomyopathy. The N-terminal PDZ domain of ZASP interacts with the C terminus of alpha-actinin-2, the major component of the Z disk, probably by forming a ternary complex with titin Z repeats. We have determined the structure of ZASP PDZ by NMR and showed that it is a classical class 1 PDZ domain that recognizes the carboxy-terminal sequence of an alpha-actinin-2 calmodulin-like domain with micromolar affinity. We also characterized the role of each component in the ternary complex ZASP/alpha-actinin-2/titin, showing that the alpha-actinin-2/ZASP PDZ interaction involves a binding surface distinct from that recognized by the titin Z repeats. ZASP PDZ structure was used to model other members of the enigma family by homology and to predict their abilities to bind alpha-actinin-2.
Organizational Affiliation:
National Institute for Medical Research, The Ridgeway, Mill Hill, London, NW7 1AA, United Kingdom.