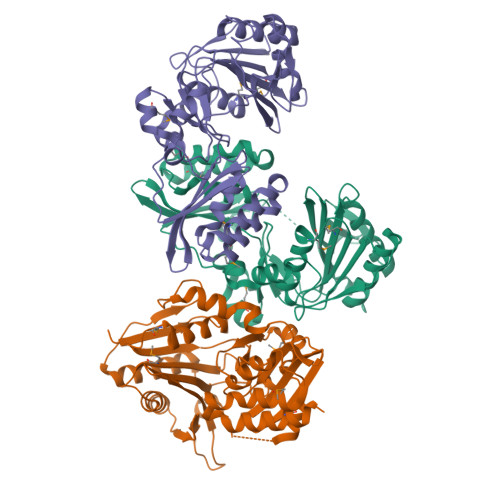
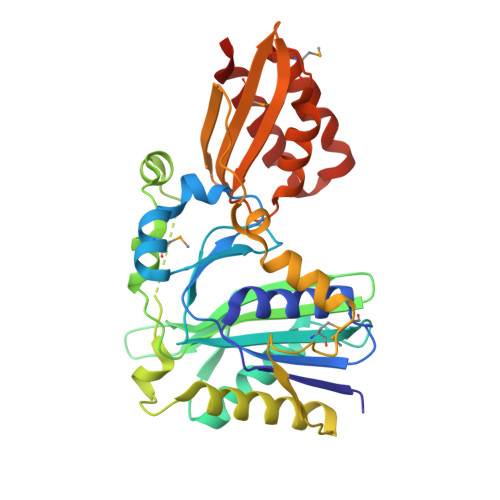
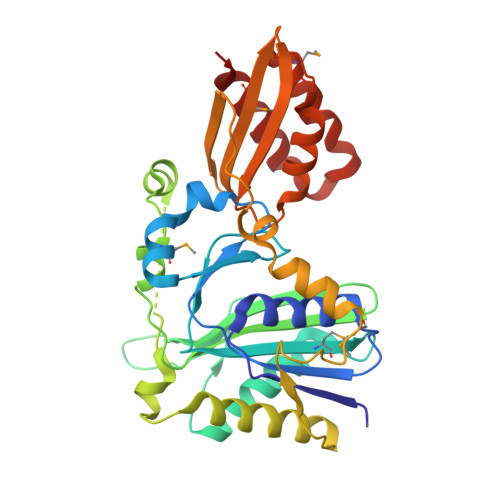
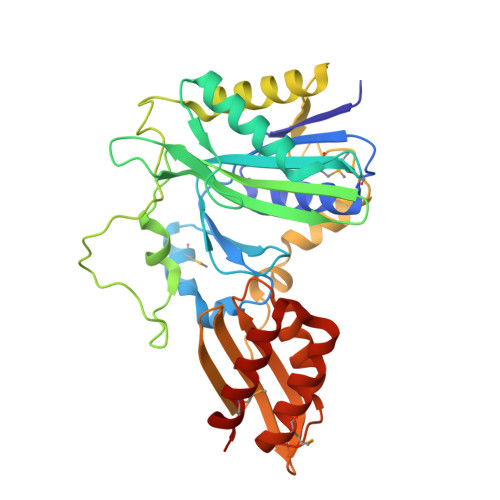
Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
Fujiwara, K., Toma, S., Okamura-Ikeda, K., Motokawa, Y., Nakagawa, A., Taniguchi, H.(2005) J Biological Chem 280: 33645-33651
- PubMed: 16043486
- DOI: https://doi.org/10.1074/jbc.M505010200
- Primary Citation of Related Structures:
1X2G, 1X2H - PubMed Abstract:
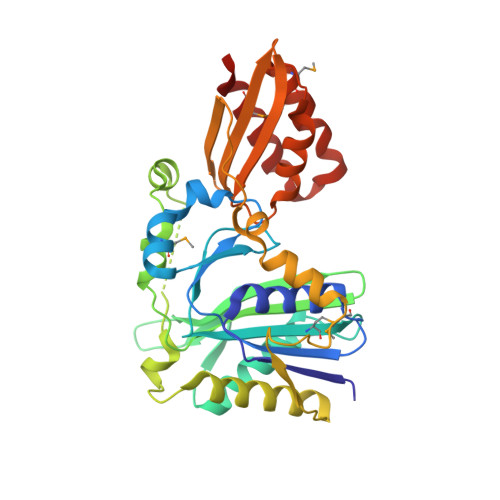
Lipoate-protein ligase A (LplA) catalyzes the formation of lipoyl-AMP from lipoate and ATP and then transfers the lipoyl moiety to a specific lysine residue on the acyltransferase subunit of alpha-ketoacid dehydrogenase complexes and on H-protein of the glycine cleavage system. The lypoyllysine arm plays a pivotal role in the complexes by shuttling the reaction intermediate and reducing equivalents between the active sites of the components of the complexes. We have determined the X-ray crystal structures of Escherichia coli LplA alone and in a complex with lipoic acid at 2.4 and 2.9 angstroms resolution, respectively. The structure of LplA consists of a large N-terminal domain and a small C-terminal domain. The structure identifies the substrate binding pocket at the interface between the two domains. Lipoic acid is bound in a hydrophobic cavity in the N-terminal domain through hydrophobic interactions and a weak hydrogen bond between carboxyl group of lipoic acid and the Ser-72 or Arg-140 residue of LplA. No large conformational change was observed in the main chain structure upon the binding of lipoic acid.
Organizational Affiliation:
Institute for Enzyme Research, the University of Tokushima, Tokushima 770-8503, Japan. fujiwara@ier.tokushima-u.ac.jp