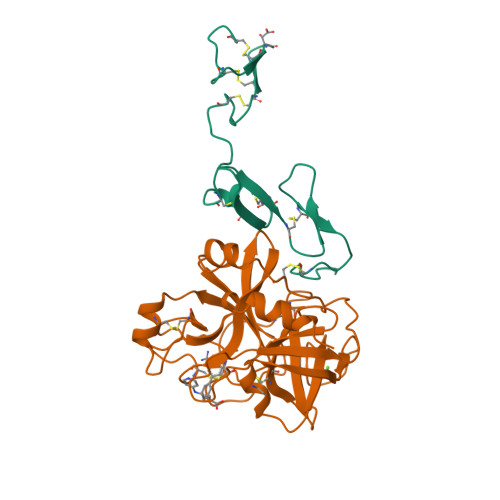
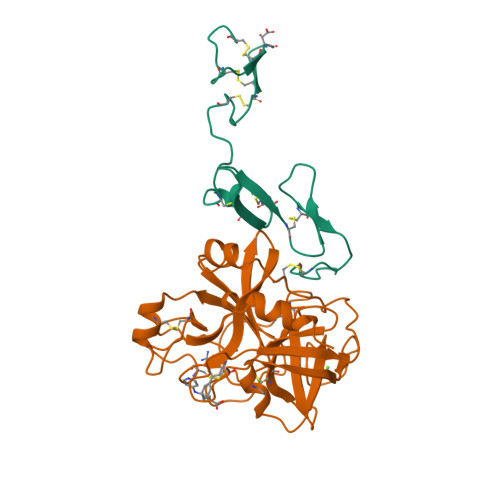
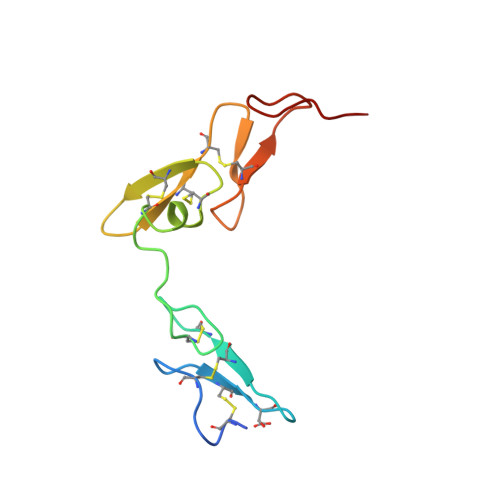
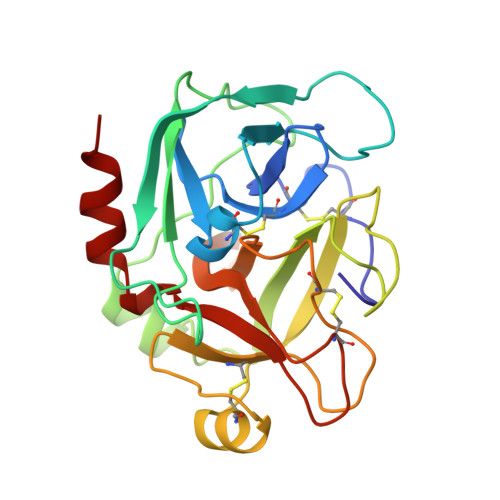
Structural basis for chemical inhibition of human blood coagulation factor Xa.
Kamata, K., Kawamoto, H., Honma, T., Iwama, T., Kim, S.H.(1998) Proc Natl Acad Sci U S A 95: 6630-6635
- PubMed: 9618463
- DOI: https://doi.org/10.1073/pnas.95.12.6630
- Primary Citation of Related Structures:
1XKA, 1XKB - PubMed Abstract:
Factor Xa, the converting enzyme of prothrombin to thrombin, has emerged as an alternative (to thrombin) target for drug discovery for thromboembolic diseases. An inhibitor has been synthesized and the crystal structure of the complex between Des[1-44] factor Xa and the inhibitor has been determined by crystallographic methods in two different crystal forms to 2.3- and 2.4-A resolution. The racemic mixture of inhibitor FX-2212, (2RS)-(3'-amidino-3-biphenylyl)-5-(4-pyridylamino)pentanoic acid, inhibits factor Xa activity by 50% at 272 nM in vitro. The S-isomer of FX-2212 (FX-2212a) was found to bind to the active site of factor Xa in both crystal forms. The biphenylamidine of FX-2212a occupies the S1-pocket, and the pyridine ring makes hydrophobic interactions with the factor Xa aryl-binding site. Several water molecules meditate inhibitor binding to residues in the active site. In contrast to the earlier crystal structures of factor Xa, such as those of apo-Des[1-45] factor Xa and Des[1-44] factor Xa in complex with a naphthyl inhibitor DX-9065a, two epidermal growth factor-like domains of factor Xa are well ordered in both our crystal forms as well as the region between the two domains, which recently was found to be the binding site of the effector cell protease receptor-1. This structure provides a basis for designing next generation inhibitors of factor Xa.
Organizational Affiliation:
Department of Chemistry and Lawrence Berkeley National Laboratory, University of California, Berkeley, CA 94720-5230, USA.