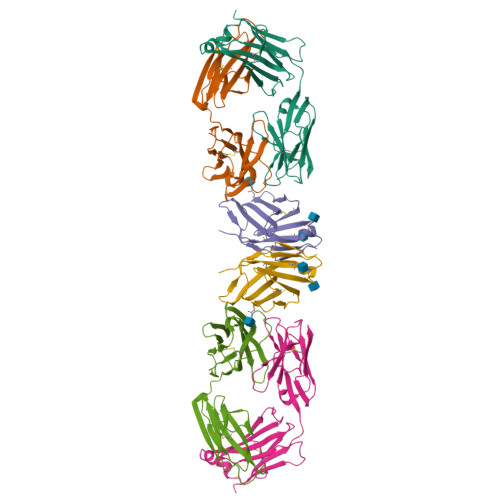
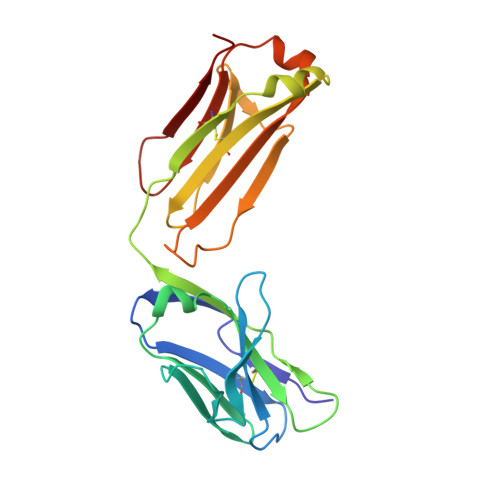
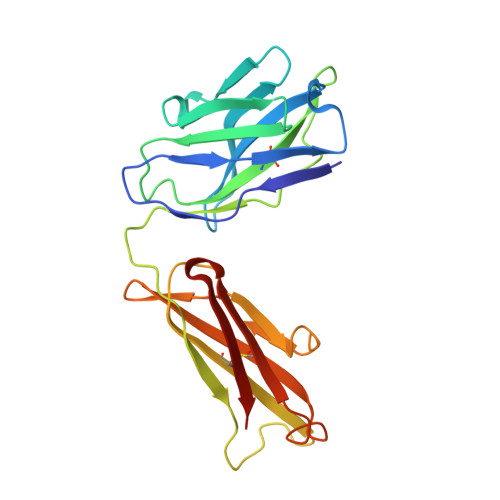
Crystal structure of a soluble CD28-Fab complex
Evans, E.J., Esnouf, R.M., Manso-Sancho, R., Gilbert, R.J.C., James, J.R., Yu, C., Fennelly, J.A., Vowles, C., Hanke, T., Walse, B., Hunig, T., Sorensen, P., Stuart, D.I., Davis, S.J.(2005) Nat Immunol 6: 271-279
- PubMed: 15696168
- DOI: https://doi.org/10.1038/ni1170
- Primary Citation of Related Structures:
1YJD - PubMed Abstract:
Naive T cell activation requires signaling by the T cell receptor and by nonclonotypic cell surface receptors. The most important costimulatory protein is the monovalent homodimer CD28, which interacts with CD80 and CD86 expressed on antigen-presenting cells. Here we present the crystal structure of a soluble form of CD28 in complex with the Fab fragment of a mitogenic antibody. Structural comparisons redefine the evolutionary relationships of CD28-related proteins, antigen receptors and adhesion molecules and account for the distinct ligand-binding and stoichiometric properties of CD28 and the related, inhibitory homodimer CTLA-4. Cryo-electron microscopy-based comparisons of complexes of CD28 with mitogenic and nonmitogenic antibodies place new constraints on models of antibody-induced receptor triggering. This work completes the initial structural characterization of the CD28-CTLA-4-CD80-CD86 signaling system.
Organizational Affiliation:
Nuffield Department of Clinical Medicine, The University of Oxford, John Radcliffe Hospital, Headington, Oxford, OX3 9DU, UK.