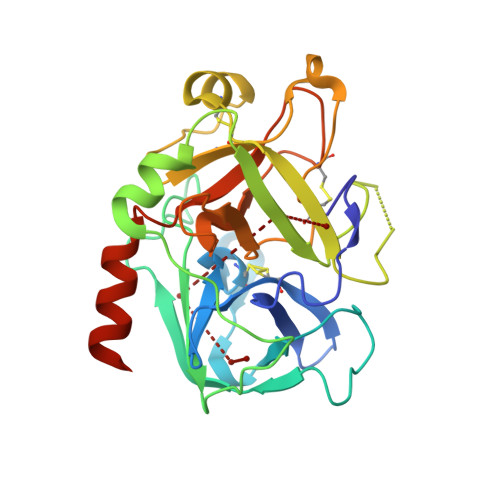
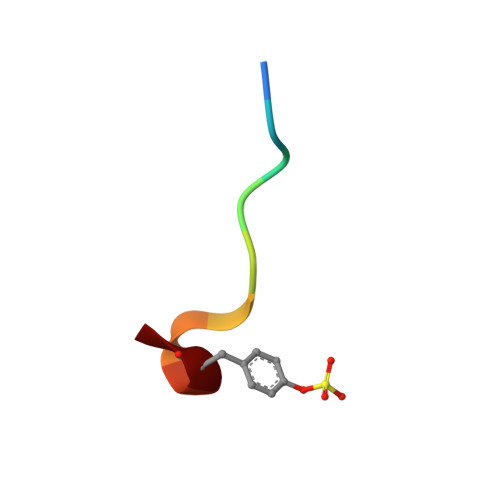
Inhibition of Human Alpha-Thrombin by a Phosphonate Tripeptide Proceeds Via a Metastable Pentacoordinated Phosphorus Intermediate
Skordalakes, E., Dodson, G.G., Green, D.S., Goodwin, C.A., Scully, M.F., Hudson, H.R., Kakkar, V.V., Deadman, J.J.(2001) J Mol Biol 311: 549
- PubMed: 11493008
- DOI: https://doi.org/10.1006/jmbi.2001.4872
- Primary Citation of Related Structures:
1H8D, 1H8I - PubMed Abstract:
X-ray crystallographic studies of human alpha-thrombin with a novel synthetic inhibitor, an acyl (alpha-aminoalkyl)phosphonate, reveal the existence of a pentacovalent phosphorus intermediate state. Crystal structures of the complex of alpha-thrombin with the phosphonate compound were determined independently using crystals of different ages. The first structure, solved from a crystal less than seven days old, showed a pentacoordinated phosphorus moiety. The second structure, determined from a crystal that was 12 weeks old, showed a tetracoordinated phosphorus moiety. In the first structure, a water molecule, made nucleophilic by coordination to His57 of alpha-thrombin, is bonded to the pentacoordinated phosphorus atom. Its position is approximately equivalent to that occupied by the water molecule responsible for hydrolytic deacylation during normal hydrolysis. The pentacoordinated phosphorus adduct collapses to give the expected pseudo tetrahedral complex, where the phosphorus atom is covalently bonded to Ser195 O(gamma). The crystallographic data presented here therefore suggest that the covalent bond formed between the inhibitor's phosphorus atom and O(gamma) of Ser195 proceeds via an addition-elimination mechanism, which involves the formation of a pentacoordinate intermediate.
Organizational Affiliation:
Chemistry Department and Biochemistry Department, Thrombosis Research Institute, Emmanuel Kaye Building, London, SW3 6LR, UK.