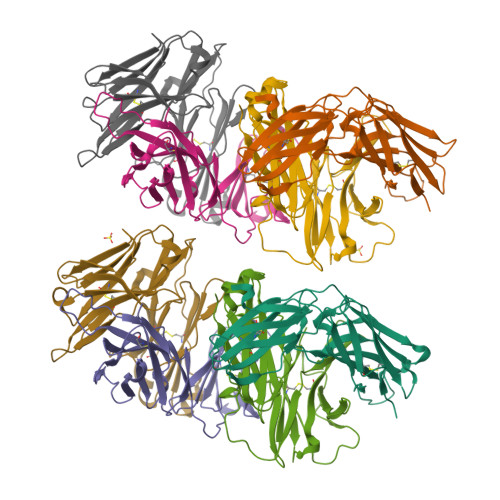
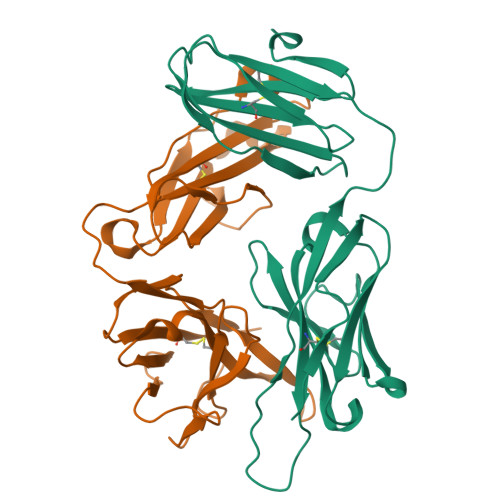
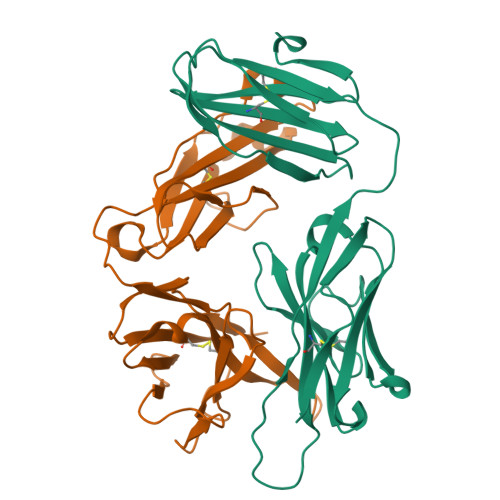
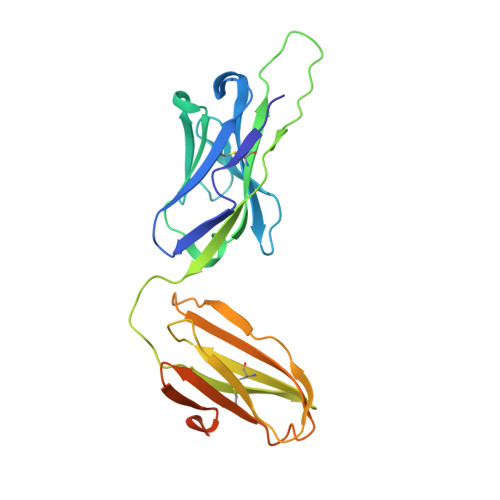
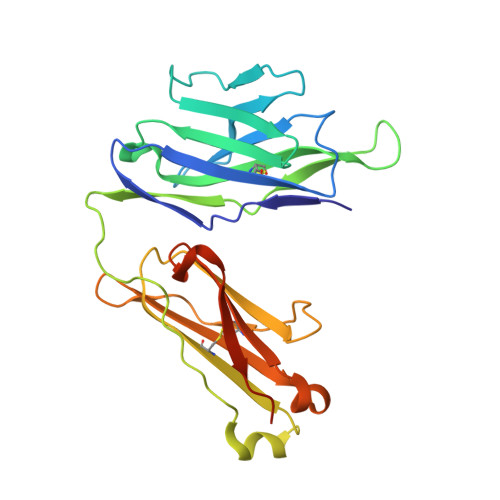
Structure of a human gammadelta T-cell antigen receptor.
Allison, T.J., Winter, C.C., Fournie, J.J., Bonneville, M., Garboczi, D.N.(2001) Nature 411: 820-824
- PubMed: 11459064
- DOI: https://doi.org/10.1038/35081115
- Primary Citation of Related Structures:
1HXM - PubMed Abstract:
T-cell antigen receptors composed of gamma and delta polypeptide chains (gammadelta TCRs) can directly recognize antigens in the form of intact proteins or non-peptide compounds, unlike alphabeta TCRs, which recognize antigens bound to major histocompatibility complex molecules (MHC). About 5% of peripheral blood T cells bear gammadelta TCRs, most of which recognize non-peptide phosphorylated antigens. Here we describe the 3.1 A resolution structure of a human gammadelta TCR from a T-cell clone that is phosphoantigen-reactive. The orientation of the variable (V) and constant (C) regions of the gammadelta TCR is unique when compared with alphabeta TCRs or antibodies, and results from an unusually small angle between the Vgamma and Cgamma domains. The complementarity-determining regions (CDRs) of the V domains exhibit a chemically reasonable binding site for phosphorylated antigens, providing a possible explanation for the canonical usage of the Vgamma9 and Vdelta2 gene segments by phosphoantigen-reactive receptors. Although the gammadelta TCR V domains are similar in overall structure to those of alphabeta TCRs, gammadelta TCR C domains are markedly different. Structural differences in Cgamma and Cdelta, and in the location of the disulphide bond between them, may enable gammadelta TCRs to form different recognition/signalling complexes than alphabeta TCRs.
Organizational Affiliation:
Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, Rockville, Maryland 20852, USA. tallison@niaid.nih.gov