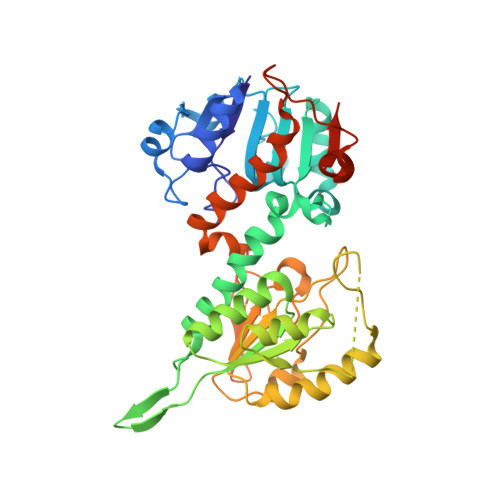
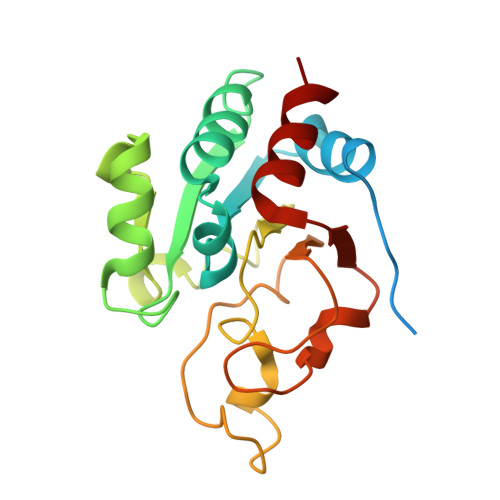
The crystal structure of an asymmetric complex of the two nucleotide binding components of proton-translocating transhydrogenase.
Cotton, N.P., White, S.A., Peake, S.J., McSweeney, S., Jackson, J.B.(2001) Structure 9: 165-176
- PubMed: 11250201
- DOI: https://doi.org/10.1016/s0969-2126(01)00571-8
- Primary Citation of Related Structures:
1HZZ - PubMed Abstract:
Membrane-bound ion translocators have important functions in biology, but their mechanisms of action are often poorly understood. Transhydrogenase, found in animal mitochondria and bacteria, links the redox reaction between NAD(H) and NADP(H) to proton translocation across a membrane. Linkage is achieved through changes in protein conformation at the nucleotide binding sites. The redox reaction takes place between two protein components located on the membrane surface: dI, which binds NAD(H), and dIII, which binds NADP(H). A third component, dII, provides a proton channel through the membrane. Intact membrane-located transhydrogenase is probably a dimer (two copies each of dI, dII, and dIII). We have solved the high-resolution crystal structure of a dI:dIII complex of transhydrogenase from Rhodospirillum rubrum-the first from a transhydrogenase of any species. It is a heterotrimer, having two polypeptides of dI and one of dIII. The dI polypeptides fold into a dimer. The loop on dIII, which binds the nicotinamide ring of NADP(H), is inserted into the NAD(H) binding cleft of one of the dI polypeptides. The cleft of the other dI is not occupied by a corresponding dIII component. The redox step in the transhydrogenase reaction is readily visualized; the NC4 atoms of the nicotinamide rings of the bound nucleotides are brought together to facilitate direct hydride transfer with A-B stereochemistry. The asymmetry of the dI:dIII complex suggests that in the intact enzyme there is an alternation of conformation at the catalytic sites associated with changes in nucleotide binding during proton translocation.
Organizational Affiliation:
School of Biosciences, University of Birmingham, Edgbaston, B15 2TT, Birmingham, United Kingdom.