Structural Basis for Low Catalytic Activity in Lys49 Phospholipases A2-A Hypothesis: The Crystal Structure of Piratoxin II Complexed to Fatty Acid
Lee, W.H., Da Silva Giotto, M.T., Marangoni, S., Toyama, M.H., Polikarpov, I., Garratt, R.C.(2001) Biochemistry 40: 28
- PubMed: 11141053
- DOI: https://doi.org/10.1021/bi0010470
- Primary Citation of Related Structures:
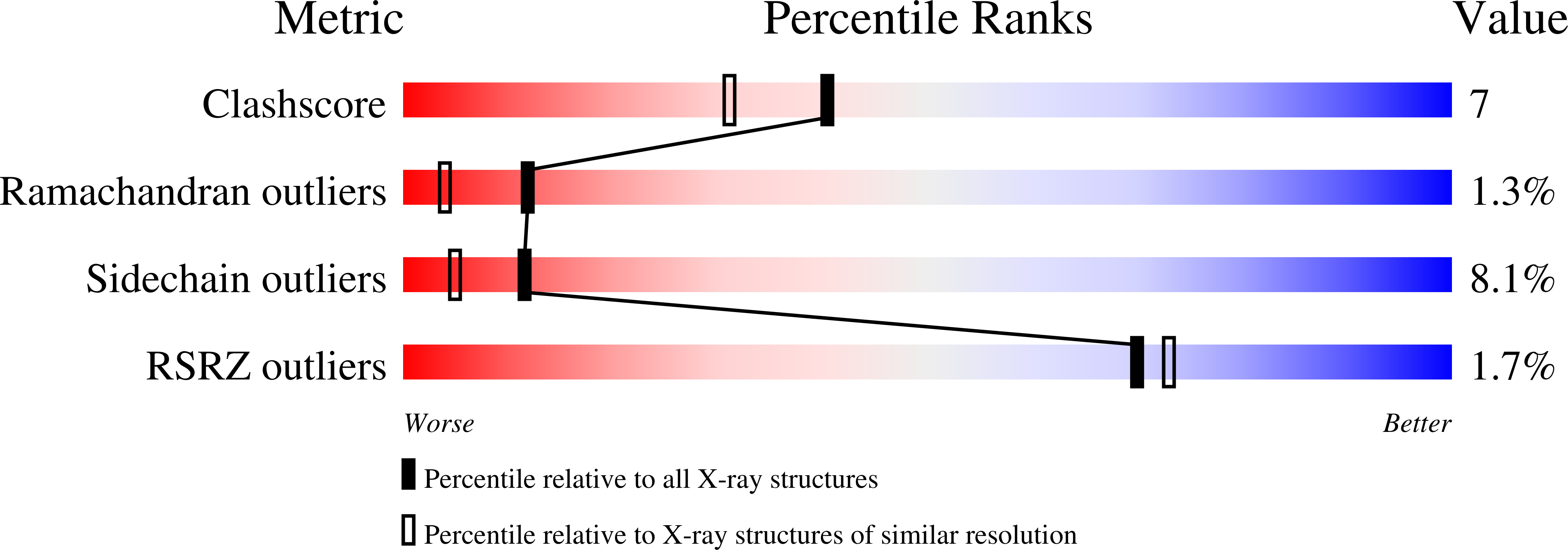
1QLL - PubMed Abstract:
Asp49 plays a fundamental role in supporting catalysis by phospholipases A2 by coordinating the calcium ion which aids in the stabilization of the tetrahedral intermediate. In several myotoxins from the venoms of Viperidae snakes, this aspartic acid is substituted by lysine. The loss of calcium binding capacity by these mutants has become regarded as the standard explanation for their greatly reduced or nonexistent phospholipolytic activity. Here we describe the crystal structure of one such Lys49 PLA2, piratoxin-II, in which a fatty acid molecule is observed within the substrate channel. This suggests that such toxins may be active enzymes in which catalysis is interrupted at the stage of substrate release. Comparison of the present structure with other PLA2s, both active and inactive, identifies Lys122 as one of the likely causes of the increased affinity for fatty acid in Lys49 enzymes. Its interaction with the mainchain carbonyl of Cys29 is expected to lead to hyperpolarization of the peptide bond between residues 29 and 30 leading to an increased affinity for the fatty acid headgroup. This strongly bound fatty acid may serve as an anchor to secure the toxin within the membrane thus facilitating its pathological effects.
Organizational Affiliation:
Laboratório Nacional de Luz Síncrotron, Caixa Postal 6192, 13083-970 Campinas, São Paulo, Brazil.