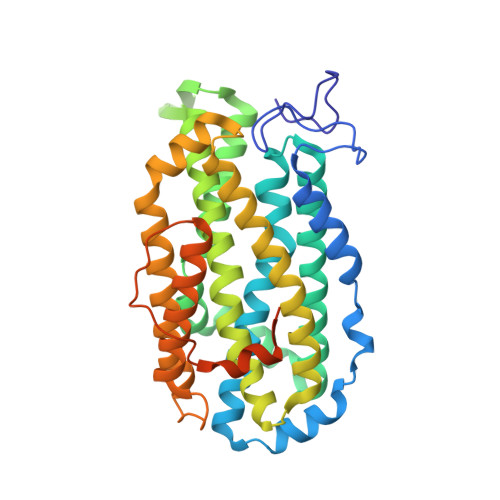
Structure and function of the Escherichia coli ribonucleotide reductase protein R2.
Nordlund, P., Eklund, H.(1993) J Mol Biol 232: 123-164
- PubMed: 8331655
- DOI: https://doi.org/10.1006/jmbi.1993.1374
- Primary Citation of Related Structures:
1RIB - PubMed Abstract:
The crystal structure of the ribonucleotide reductase free radical protein R2 from Escherichia coli has been determined by multiple isomorphous replacement and twofold molecular averaging. The structure has been refined at 2.2 A resolution to R = 0.175. The subunit structure of the R2 protein has a novel fold where the basic motif is a bundle of eight long helices. The R2 dimer has two equivalent dinuclear iron centers. Each iron center is well buried in the subunit. The iron atoms have both histidine and carboxyl acid ligands and are bridged by an oxide ion and the carboxylate group of Glu115. One iron atom is octahedrally coordinated with small deviations from ideal values, while the coordination of the other iron ion is more distorted, mainly due to the fact that Asp84 is a bidental ligand to this iron atom. The oxidation of the enzymatically essential tyrosine residue (Tyr122) and the dinuclear iron center by molecular oxygen is suggested to take part in a suitable conserved oxygen-binding pocket between the iron center and the tyrosine zeta-oxygen 5.3 A away from the closest iron ion. The tyrosine proton can be abstracted by the dioxygen and the deprotonated tyrosine residue is then more easily oxidized to a radical species. Tyr122 is buried inside the protein about 10 A from the surface. This has the consequence that the tyrosyl radical cannot participate directly in hydrogen abstraction from the substrate ribose at the active site of the holoenzyme located on the R1 subunit. The radical must then be indirectly involved in the mechanism of the enzyme and an electron transfer reaction between the active site and the tyrosine must take place. Based on the analysis of the available ribonucleotide reductase sequences, the binding surface for the large ribonucleotide reductase protein R1, and a possible route for an electron transport between the buried radical and this surface is described.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Biomedical Center, Uppsala.