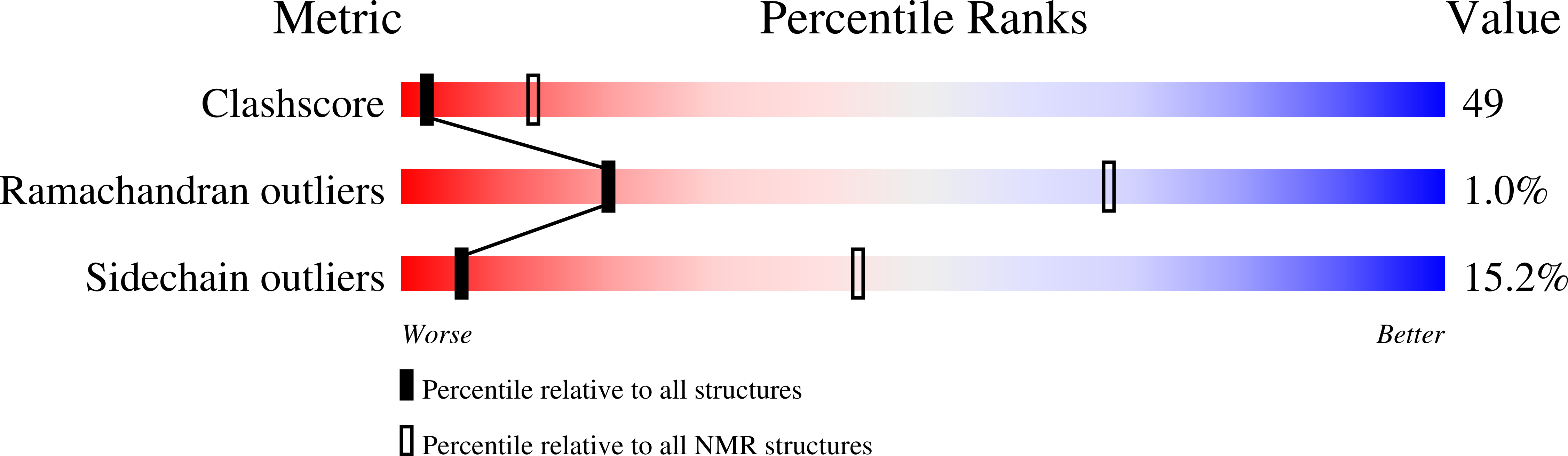Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type.
Skelton, N.J., Aspiras, F., Ogez, J., Schall, T.J.(1995) Biochemistry 34: 5329-5342
- PubMed: 7537088
- DOI: https://doi.org/10.1021/bi00016a004
- Primary Citation of Related Structures:
1RTN, 1RTO - PubMed Abstract:
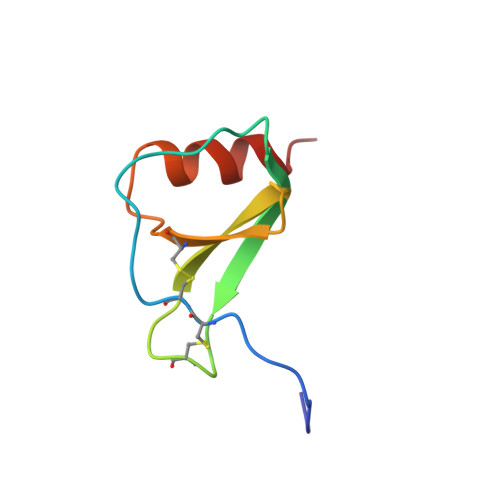
1H NMR has been used to investigate the structural properties of RANTES, a protein from the C-C branch of the chemotactic cytokine family that has a strong chemoattractive effect on monocytes, lymphocytes, and eosinophils. Titration of pH from 5.0 to 2.5 indicates that RANTES is extensively aggregated in solution above pH 4.0. At pH 3.7 the protein is mostly dimeric, although this species does dissociate to the monomer with a Kd of 35 microM. NMR data have been acquired and resonance assignments made for the dimeric species. Structures of the dimer have been generated by distance geometry and simulated annealing calculations that utilized 1956 intramolecular distance restraints, 120 intermolecular distance restraints, 164 dihedral angle restraints, and 68 restraints enforcing 34 hydrogen bonds (17.0 restraints per residue). The structure is well-defined (average root mean square deviation from the average structure of 0.38 +/- 0.06 and 0.53 +/- 0.12 A for backbone heavy atoms of residues 4-66 of the monomer and dimer, respectively). Each monomer consists of a C-terminal alpha-helix packing against a three-stranded antiparallel beta-sheet and two short N-terminal beta-strands; dimerization occurs between the N-terminal regions of each monomer. This quaternary structure is very different from that of the C-X-C chemokines such as interleukin-8 and melanoma growth stimulatory activity but similar to that found for the C-C chemokine macrophage inflammatory factor 1 beta. Distinct structural differences between RANTES and other chemokines at both the tertiary and quaternary level are discussed with regard to the distinct biological functions of the C-C and C-X-C members of this protein family.
Organizational Affiliation:
Department of Protein Engineering, Genetech, Inc., South San Francisco, California 94080, USA.