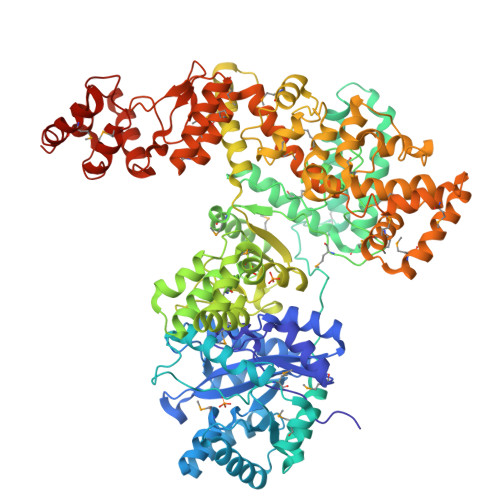
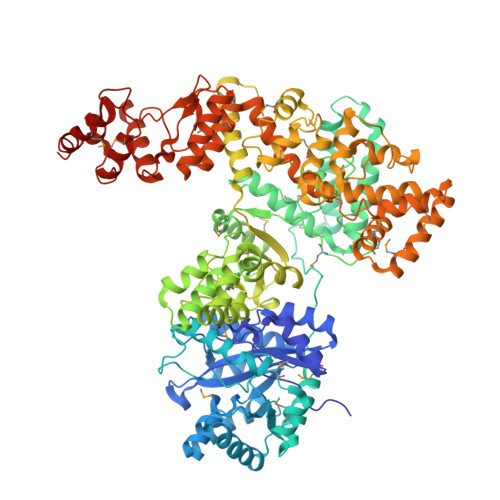
Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Lamers, M.H., Georgescu, R.E., Lee, S.G., O'Donnell, M., Kuriyan, J.(2006) Cell 126: 881-892
- PubMed: 16959568
- DOI: https://doi.org/10.1016/j.cell.2006.07.028
- Primary Citation of Related Structures:
2HNH, 2HQA - PubMed Abstract:
Bacterial replicative DNA polymerases such as Polymerase III (Pol III) share no sequence similarity with other polymerases. The crystal structure, determined at 2.3 A resolution, of a large fragment of Pol III (residues 1-917), reveals a unique chain fold with localized similarity in the catalytic domain to DNA polymerase beta and related nucleotidyltransferases. The structure of Pol III is strikingly different from those of members of the canonical DNA polymerase families, which include eukaryotic replicative polymerases, suggesting that the DNA replication machinery in bacteria arose independently. A structural element near the active site in Pol III that is not present in nucleotidyltransferases but which resembles an element at the active sites of some canonical DNA polymerases suggests that, at a more distant level, all DNA polymerases may share a common ancestor. The structure also suggests a model for interaction of Pol III with the sliding clamp and DNA.
Organizational Affiliation:
Howard Hughes Medical Institute, Department of Molecular and Cell Biology and Department of Chemistry, University of California, Berkeley, CA 94720, USA.