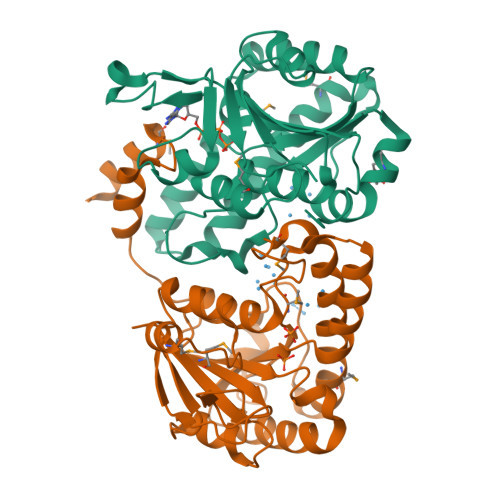
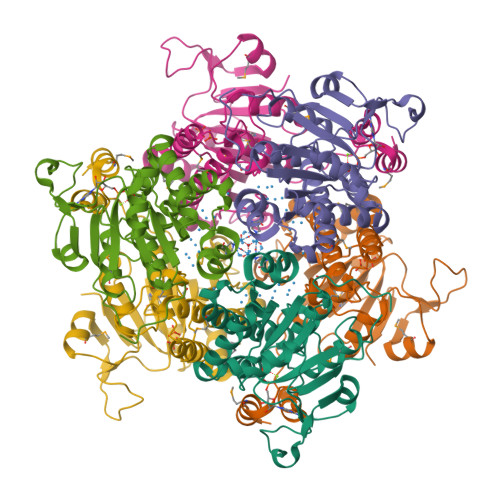
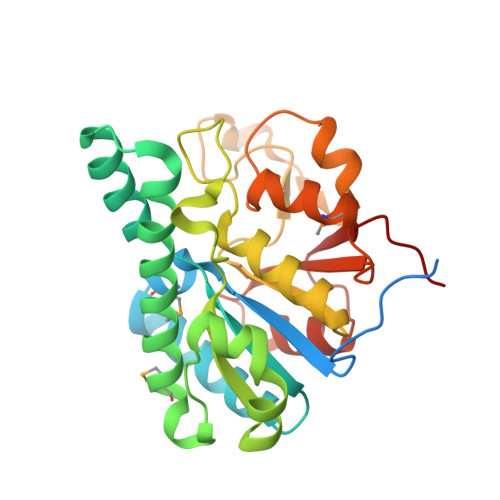
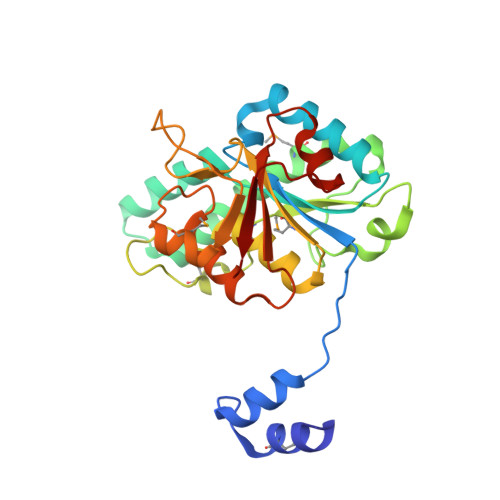
Towards Biological Supramolecular Chemistry: A Variety of Pocket-Templated, Individual Metal Oxide Cluster Nucleations in the Cavity of a Mo/W-Storage Protein.
Schemberg, J., Schneider, K., Demmer, U., Warkentin, E., Muller, A., Ermler, U.(2007) Angew Chem Int Ed Engl 46: 2408-2413
- PubMed: 17304608
- DOI: https://doi.org/10.1002/anie.200604858
- Primary Citation of Related Structures:
2OGX
Organizational Affiliation:
Fakultät für Chemie der Universität, Postfach 100131, 33501 Bielefeld, Germany.